bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-3_CDS_annotation_glimmer3.pl_2_8
Length=354
Score E
Sequences producing significant alignments: (Bits) Value
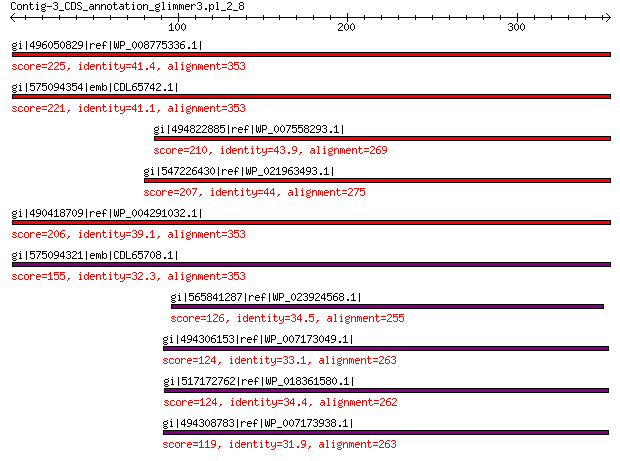
gi|496050829|ref|WP_008775336.1| hypothetical protein 225 2e-64
gi|575094354|emb|CDL65742.1| unnamed protein product 221 9e-63
gi|494822885|ref|WP_007558293.1| hypothetical protein 210 2e-58
gi|547226430|ref|WP_021963493.1| putative uncharacterized protein 207 1e-57
gi|490418709|ref|WP_004291032.1| hypothetical protein 206 4e-57
gi|575094321|emb|CDL65708.1| unnamed protein product 155 6e-39
gi|565841287|ref|WP_023924568.1| hypothetical protein 126 2e-28
gi|494306153|ref|WP_007173049.1| hypothetical protein 124 3e-28
gi|517172762|ref|WP_018361580.1| hypothetical protein 124 4e-28
gi|494308783|ref|WP_007173938.1| hypothetical protein 119 1e-26
>gi|496050829|ref|WP_008775336.1| hypothetical protein [Bacteroides sp. 2_2_4]
gi|229448893|gb|EEO54684.1| putative capsid protein (F protein) [Bacteroides sp. 2_2_4]
Length=580
Score = 225 bits (573), Expect = 2e-64, Method: Compositional matrix adjust.
Identities = 146/357 (41%), Positives = 199/357 (56%), Gaps = 42/357 (12%)
Query 2 GILPNSQFGDLAVVDLGTISASGSKIPVGAYDGVNDSGTFKQFQTSSESYNGSSDKTTQI 61
G+LP Q+GD A V++ + ++ V DG + GS +T +
Sbjct 262 GVLPRQQYGDTAAVNVNLSNVLSAQYMVQTPDG--------------DPVGGSPFSSTGV 307
Query 62 YPRNPDSISVRADSNLWAVLGESSDLKLKFSVLALRQAEALQKWKEITQSVDTNYRDQIK 121
NL V G + F+VLALRQAE LQKWKEITQS + +Y+DQI+
Sbjct 308 --------------NLQTVNGSGT-----FTVLALRQAEFLQKWKEITQSGNKDYKDQIE 348
Query 122 AHFGVSVPQSEAHMAKYIGGVARNLDISEVVNNFLPGPGNDSSQAYIYGKGVGSGQGSMT 181
H+ VSV ++ + M+ Y+GG +LDI+EVVNN + G S+ A I GKGV G G ++
Sbjct 349 KHWNVSVGEAYSEMSLYLGGTTASLDINEVVNNNITG----SNAADIAGKGVVVGNGRIS 404
Query 182 FNTGSGYYILMCIYHAVPLLDYSISGPDGQNLVTSVEDLPIPEFDNIGMESVPAVELMNS 241
F+ G Y ++MCIYH++PLLDY+ + + D IPEFD +GMESVP V LMN
Sbjct 405 FDAGERYGLIMCIYHSLPLLDYTTDLVNPAFTKINSTDFAIPEFDRVGMESVPLVSLMN- 463
Query 242 SLYSNVNSADKILGYSPRYYNWKTKIDRIHGAFTTTLKDWVAPIDD-SFLYSL-FGDSLS 299
L S+ N ILGY+PRY ++KT +D GAF TTLK WV D+ S + L + D +
Sbjct 464 PLQSSYNVGSSILGYAPRYISYKTDVDSSVGAFKTTLKSWVMSYDNQSVINQLNYQDDPN 523
Query 300 TYKG--VTWPFFKVNPNTLDDLFAVKVDSTWETDQLLVNCNVGCYVTRPLSADGVPY 354
G V + FKVNPN +D LFAV ++ +TDQ L + V R L DG+PY
Sbjct 524 NSPGTLVNYTNFKVNPNCVDPLFAVAASNSIDTDQFLCSSFFDVKVVRNLDTDGLPY 580
>gi|575094354|emb|CDL65742.1| unnamed protein product [uncultured bacterium]
Length=615
Score = 221 bits (564), Expect = 9e-63, Method: Compositional matrix adjust.
Identities = 145/387 (37%), Positives = 205/387 (53%), Gaps = 51/387 (13%)
Query 2 GILPNSQFGDLAVV------------DLGTISASGSKIP---------VGAYDGVNDSGT 40
G+LP +Q+G +VV D G I + + P VG GV++
Sbjct 246 GVLPVAQYGSASVVPINGQLNVISNGDSGPIFKTSTPDPGTPGTSYVTVGGNIGVDN--- 302
Query 41 FKQFQTSSESYN--GSSDKTTQIYPRNPDSISVRADSNLWAVLGESSDLKLK------FS 92
+ F S + N S+D + +P N + S+ LW E+ +L ++
Sbjct 303 -RSFGVSGSTLNVGKSADPSGYGFPSNASTRSL-----LW----ENPNLIIENNQGFYVP 352
Query 93 VLALRQAEALQKWKEITQSVDTNYRDQIKAHFGVSVPQSEAHMAKYIGGVARNLDISEVV 152
+LALRQAE LQKWKE++ S + +Y+ QI+ H+G+ V +H A+Y+GG A +LDI+EV+
Sbjct 353 ILALRQAEFLQKWKEVSVSGEEDYKSQIEKHWGIKVSDFLSHQARYLGGCATSLDINEVI 412
Query 153 NNFLPGPGNDSSQAYIYGKGVGSGQGSMTFNTGSGYYILMCIYHAVPLLDYSISGPDGQN 212
NN + G + A I GKG +G GS+ F + Y I+MCIYH +P++DY SG D
Sbjct 413 NNNITG----DNAADIAGKGTFTGNGSIRFESKGEYGIIMCIYHVLPIVDYVGSGVDHSC 468
Query 213 LVTSVEDLPIPEFDNIGMESVPAVELMNSSLYSNVNSADKILGYSPRYYNWKTKIDRIHG 272
+ PIPE D IGMESVP V MN S+ SAD LGY+PRY +WKT +DR G
Sbjct 469 TLVDATSFPIPELDQIGMESVPLVRAMNPVKESDTPSADTFLGYAPRYIDWKTSVDRSVG 528
Query 273 AFTTTLKDWVAPIDDSFLYSLFGDSLSTYKGV-----TWPFFKVNPNTLDDLFAVKVDST 327
F +L+ W P+ D L S + + V FFKVNP+ +D LFAV DST
Sbjct 529 DFADSLRTWCLPVGDKELTSANSLNFPSNPNVEPDSIAAGFFKVNPSIVDPLFAVVADST 588
Query 328 WETDQLLVNCNVGCYVTRPLSADGVPY 354
+TD+ L + V R L +G+PY
Sbjct 589 VKTDEFLCSSFFDVKVVRNLDVNGLPY 615
>gi|494822885|ref|WP_007558293.1| hypothetical protein [Bacteroides plebeius]
gi|198272099|gb|EDY96368.1| putative capsid protein (F protein) [Bacteroides plebeius DSM
17135]
Length=613
Score = 210 bits (535), Expect = 2e-58, Method: Compositional matrix adjust.
Identities = 118/279 (42%), Positives = 163/279 (58%), Gaps = 16/279 (6%)
Query 86 DLKLKFSVLALRQAEALQKWKEITQSVDTNYRDQIKAHFGVSVPQSEAHMAKYIGGVARN 145
D S+LALR+AEA QKWKE+ + + +Y QI+AH+G SV ++ + M +++G + +
Sbjct 341 DSSFGVSILALRRAEAAQKWKEVALASEEDYPSQIEAHWGQSVNKAYSDMCQWLGSINID 400
Query 146 LDISEVVNNFLPGPGNDSSQAYIYGKGVGSGQGSMTFNTGSGYYILMCIYHAVPLLDYSI 205
L I+EVVNN + G + A I GKG SG GS+ FN G Y I+MC++H +P LDY
Sbjct 401 LSINEVVNNNITG----ENAADIAGKGTMSGNGSINFNVGGQYGIVMCVFHVLPQLDYIT 456
Query 206 SGPDGQNLVTSVEDLPIPEFDNIGMESVPAVELMNSSLYSNVN---SADKILGYSPRYYN 262
S P +T+V D PIPEFD IGME VP + +N + + S + GY+P+YYN
Sbjct 457 SAPHFGTTLTNVLDFPIPEFDKIGMEQVPVIRGLNPVKPKDGDFKVSPNLYFGYAPQYYN 516
Query 263 WKTKIDRIHGAFTTTLKDWVAPIDDSFLYSLFGDSLS-------TYKGVTWPFFKVNPNT 315
WKT +D+ G F +LK W+ P DD L L DS+ V FFKV+P+
Sbjct 517 WKTTLDKSMGEFRRSLKTWIIPFDDEAL--LAADSVDFPDNPNVEADSVKAGFFKVSPSV 574
Query 316 LDDLFAVKVDSTWETDQLLVNCNVGCYVTRPLSADGVPY 354
LD+LFAVK +S TDQ L + V R L +G+PY
Sbjct 575 LDNLFAVKANSDLNTDQFLCSTLFDVNVVRSLDPNGLPY 613
>gi|547226430|ref|WP_021963493.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
gi|524103382|emb|CCY83994.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
Length=573
Score = 207 bits (527), Expect = 1e-57, Method: Compositional matrix adjust.
Identities = 121/280 (43%), Positives = 169/280 (60%), Gaps = 11/280 (4%)
Query 80 VLGESSDLKLKFSVLALRQAEALQKWKEITQSVDTNYRDQIKAHFGVSVPQSEAHMAKYI 139
V+ +S+ SVLALRQAE LQKW+EI QS +Y+ Q++ HF VS + + KY+
Sbjct 300 VVNNNSNTTAGLSVLALRQAECLQKWREIAQSGKMDYQTQMQKHFNVSPSATLSGHCKYL 359
Query 140 GGVARNLDISEVVNNFLPGPGNDSSQAYIYGKGVGSGQGSMTFNTGSGYYILMCIYHAVP 199
GG NLDISEVVN L G +QA I GKG G+ G+ S + I+MCIYH +P
Sbjct 360 GGWTSNLDISEVVNTNLTG----DNQADIQGKGTGTLNGNKVDFESSEHGIIMCIYHCLP 415
Query 200 LLDYSISGPDGQNLVTSVEDLPIPEFDNIGMESVPAVELMN--SSLYSNVNSADKILGYS 257
LLD+SI+ QN T+ D IPEFD++GM+ + E++ L S+ +S + +GY
Sbjct 416 LLDWSINRIARQNFKTTFTDYAIPEFDSVGMQQLYPSEMIFGLEDLPSDPSSIN--MGYV 473
Query 258 PRYYNWKTKIDRIHGAFTTTLKDWVAPIDDSFLYSL---FGDSLSTYKGVTWPFFKVNPN 314
PRY + KT ID IHG+F TL WV+P+ DS++ + D+ + +T+ FFKVNP+
Sbjct 474 PRYADLKTSIDEIHGSFIDTLVSWVSPLTDSYISAYRQACKDAGFSDITMTYNFFKVNPH 533
Query 315 TLDDLFAVKVDSTWETDQLLVNCNVGCYVTRPLSADGVPY 354
+D++F VK DST TDQLL+N R +G+PY
Sbjct 534 IVDNIFGVKADSTINTDQLLINSYFDIKAVRNFDYNGLPY 573
>gi|490418709|ref|WP_004291032.1| hypothetical protein [Bacteroides eggerthii]
gi|217986636|gb|EEC52970.1| putative capsid protein (F protein) [Bacteroides eggerthii DSM
20697]
Length=578
Score = 206 bits (523), Expect = 4e-57, Method: Compositional matrix adjust.
Identities = 138/374 (37%), Positives = 188/374 (50%), Gaps = 70/374 (19%)
Query 2 GILPNSQFGDLAVVDLGTISASGSKIPVGAYDGVNDSGTFKQFQTSSESYNGSSDKTTQI 61
G+LP+ Q+G+ AV + T +G T F T S
Sbjct 254 GVLPHQQYGETAVASI-TPDVTGKL-------------TLSNFSTVGTS----------- 288
Query 62 YPRNPDSISVRADSNLWAVLGESSDLKLKFSVLALRQAEALQKWKEITQSVDTNYRDQIK 121
P + S A NL A D S+L LRQAE LQKWKEITQS + +Y+DQ++
Sbjct 289 ----PTTASGTATKNLPAF-----DTVGDLSILVLRQAEFLQKWKEITQSGNKDYKDQLE 339
Query 122 AHFGVSVPQSEAHMAKYIGGVARNLDISEVVNNFLPGPGNDSSQAYIYGKGVGSGQGSMT 181
H+GVSV + + Y+GGV+ ++DI+EV+N + G S+ A I GKGVG G +
Sbjct 340 KHWGVSVGDGFSELCTYLGGVSSSIDINEVINTNITG----SAAADIAGKGVGVANGEIN 395
Query 182 FNTGSGYYILMCIYHAVPLLDYSISGPDGQNLVTSVEDLPIPEFDNIGMESVPAVELMNS 241
FN+ Y ++MCIYH +PLLDY+ D L + D IPEFD +GM+S+P V+LMN
Sbjct 396 FNSNGRYGLIMCIYHCLPLLDYTTDMLDPAFLKVNSTDYAIPEFDRVGMQSMPLVQLMN- 454
Query 242 SLYSNVNSADKILGYSPRYYNWKTKIDRIHGAFTTTLKDWVAPIDDSFLYSLFGDSLSTY 301
L S N++ +LGY PRY ++KT +D+ G F TL WV +G+ +S
Sbjct 455 PLRSFANASGLVLGYVPRYIDYKTSVDQSVGGFKRTLNSWVIS---------YGN-ISVL 504
Query 302 KGVTWP---------------------FFKVNPNTLDDLFAVKVDSTWETDQLLVNCNVG 340
K VT P FFKVNP+ LD +FAV+ TDQ L +
Sbjct 505 KQVTLPNDAPPIEPSEPVPSVAPMNFTFFKVNPDCLDPIFAVQAGDDTNTDQFLCSSFFD 564
Query 341 CYVTRPLSADGVPY 354
R L DG+PY
Sbjct 565 IKAVRNLDTDGLPY 578
>gi|575094321|emb|CDL65708.1| unnamed protein product [uncultured bacterium]
Length=642
Score = 155 bits (393), Expect = 6e-39, Method: Compositional matrix adjust.
Identities = 114/367 (31%), Positives = 180/367 (49%), Gaps = 26/367 (7%)
Query 2 GILPNSQFGDLAVVDLGTISASGSKIPVGAYDGVNDSGTFKQFQTSSE-SYNGSSDKTTQ 60
G+LP SQFG +VV+L +ASGS + G DSG ++ E +S
Sbjct 286 GVLPTSQFGSESVVNLNLGNASGSAVLNGTTS--KDSGRWRTTTGEWEMEQRVASSANGN 343
Query 61 IYPRNPDSISVRADSNLWAVLGESSDLKLKFSVLALRQAEALQKWKEITQSVDTNYRDQI 120
+ N + + D + ++ L S++ALR A A QK+KEI + D +++ Q+
Sbjct 344 LKLDNSNGTFISHDHTFSGNVAINTSLSGNLSIIALRNALAAQKYKEIQLANDVDFQSQV 403
Query 121 KAHFGVSVPQSEAHMAKYIGGVARNLDISEVVNNFLPGPGNDSSQAYIYGKG-VGSGQGS 179
+AHFG+ P + + +IGG + ++I+E +N L G + YG G+G S
Sbjct 404 EAHFGIK-PDEKNENSLFIGGSSSMININEQINQNLSGDNKAT-----YGAAPQGNGSAS 457
Query 180 MTFNTGSGYYILMCIYHAVPLLDYSISGPDGQNLVTSVEDLPIPEFDNIGMESVPAVELM 239
+ F T Y +++ IY P+LD++ G D T D IPE D+IGM+ E+
Sbjct 458 IKF-TAKTYGVVIGIYRCTPVLDFAHLGIDRTLFKTDASDFVIPEMDSIGMQQTFRCEVA 516
Query 240 NSSLYSNVNSADKI-----------LGYSPRYYNWKTKIDRIHGAFTTTLKDWVAPIDDS 288
+ Y++ A ++ GY+PRY +KT DR +GAF +LK WV I+
Sbjct 517 APAPYNDEFKAFRVGDGSSPDMSETYGYAPRYSEFKTSYDRYNGAFCHSLKSWVTGIN-- 574
Query 289 FLYSLFGDSLSTYKGVTWP-FFKVNPNTLDDLFAVKVDSTWETDQLLVNCNVGCYVTRPL 347
++ + +T+ G+ P F P+ + +LF V + + DQL V CY TR L
Sbjct 575 -FDAIQNNVWNTWAGINAPNMFACRPDIVKNLFLVSSTNNSDDDQLYVGMVNMCYATRNL 633
Query 348 SADGVPY 354
S G+PY
Sbjct 634 SRYGLPY 640
>gi|565841287|ref|WP_023924568.1| hypothetical protein [Prevotella nigrescens]
gi|564729907|gb|ETD29851.1| hypothetical protein HMPREF1173_00033 [Prevotella nigrescens
CC14M]
Length=656
Score = 126 bits (316), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 88/267 (33%), Positives = 133/267 (50%), Gaps = 23/267 (9%)
Query 96 LRQAEALQKWKEITQSVD-TNYRDQIKAHFGVSVPQSEAHMAKYIGGVARNLDISEVVNN 154
+R AL+K E T++ + +Y +QI AHFG VP+S + A +IGG + ISEVV
Sbjct 396 IRAMFALEKMLERTRAANGLDYSNQIAAHFGFKVPESRKNCASFIGGFDNQISISEVVTT 455
Query 155 FLPGPGNDSSQAYIYGKGVGSGQGSMTFNTGSGYY------ILMCIYHAVPLLDYSISGP 208
+S + G+ G G G+M N+G Y ++MCIY P +DY
Sbjct 456 SNGSVDGTASTGSVVGQVFGKGIGAM--NSGHISYDVKEHGLIMCIYSIAPQVDYDAREL 513
Query 209 DGQNLVTSVEDLPIPEFDNIGMESVPAVEL---MNSSLYSNVNSADKILGYSPRYYNWKT 265
D N S ED PEF+N+GM+ V +L +NS+ + + + +LGYS RY +KT
Sbjct 514 DPFNRKFSREDYFQPEFENLGMQPVIQSDLCLCINSAKSDSSDQHNNVLGYSARYLEYKT 573
Query 266 KIDRIHGAFTT--TLKDWVAPIDDSFLYSLFGDSLSTYKGVTWPFFKVNPNTLDDLFAVK 323
D I G F + +L W P ++ Y+ + ++ P V+P L+ +FAVK
Sbjct 574 ARDIIFGEFMSGGSLSAWATPKNN---YTF------EFGKLSLPDLLVDPKVLEPIFAVK 624
Query 324 VDSTWETDQLLVNCNVGCYVTRPLSAD 350
+ + TDQ LVN RP+ +
Sbjct 625 YNGSMSTDQFLVNSYFDVKAIRPMQVN 651
>gi|494306153|ref|WP_007173049.1| hypothetical protein [Prevotella bergensis]
gi|270333881|gb|EFA44667.1| putative capsid protein (F protein) [Prevotella bergensis DSM
17361]
Length=519
Score = 124 bits (310), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 87/275 (32%), Positives = 129/275 (47%), Gaps = 31/275 (11%)
Query 91 FSVLALRQAEALQKWKEITQSVDTNYRDQIKAHFGVSVPQSEAHMAKYIGGVARNLDISE 150
FSV +LR A A+ K +T ++DQ++AH+GV +P S Y+GG +L +S+
Sbjct 262 FSVSSLRSAFAVDKLLSVTMRAGKTFQDQMRAHYGVEIPDSRDGRVNYLGGFDSDLQVSD 321
Query 151 VVN-------NFLPGPGNDSSQAYIYGKGVGSGQGSMTFNTGSGYYILMCIYHAVPLLDY 203
V + P G I GKG GSG+G + F+ + +LMCIY VP + Y
Sbjct 322 VTQTSGTTATEYKPEAG---YLGRIAGKGTGSGRGRIVFDAKE-HGVLMCIYSLVPQIQY 377
Query 204 SISGPDGQNLVTSVEDLPIPEFDNIGMESVPAVELMNSSLYSNVNSAD---KILGYSPRY 260
+ D D PEF+N+GM+ +NSS S+ + D +LGY PRY
Sbjct 378 DCTRLDPMVDKLDRFDFFTPEFENLGMQP------LNSSYISSFCTPDPKNPVLGYQPRY 431
Query 261 YNWKTKIDRIHGAFTT--TLKDWVAPIDDSFLYSLFGDSLSTYKGVTWPFFKVNPNTLDD 318
+KT +D HG F L W S F +T+ + FK++P L+
Sbjct 432 SEYKTALDINHGQFAQNDALSSWSV--------SRFR-RWTTFPQLEIADFKIDPGCLNS 482
Query 319 LFAVKVDSTWETDQLLVNCNVGCYVTRPLSADGVP 353
+F V+ + T TD + CN +S DG+P
Sbjct 483 VFPVEFNGTESTDCVFGGCNFNIVKVSDMSVDGMP 517
>gi|517172762|ref|WP_018361580.1| hypothetical protein [Prevotella nanceiensis]
Length=568
Score = 124 bits (311), Expect = 4e-28, Method: Compositional matrix adjust.
Identities = 90/275 (33%), Positives = 134/275 (49%), Gaps = 26/275 (9%)
Query 92 SVLALRQAEALQKWKEITQSVDTNYRDQIKAHFGVSVPQSEAHMAKYIGGVARNLDISEV 151
SV +R A AL+K +T Y++Q++AHFG+SV + YIGG N+ + +V
Sbjct 305 SVADIRNAFALEKLASVTMRAGKTYKEQMEAHFGISVEEGRDGRCTYIGGFDSNIQVGDV 364
Query 152 VNN---FLPGPGNDSSQAYI---YGKGVGSGQGSMTFNTGSGYYILMCIYHAVPLLDYSI 205
+ + G + S Y+ GK GSG G + F+ + ILMCIY VP + Y
Sbjct 365 TQSSGTTVTGTKDTSFGGYLGRTTGKATGSGSGHIRFDAKE-HGILMCIYSLVPDVQYDS 423
Query 206 SGPDGQNLVTSVE--DLPIPEFDNIGMESVPAVELMNSSLYSNVNSADKI-----LGYSP 258
D V +E D +PEF+N+GM+ + A + S Y+N + +I G+ P
Sbjct 424 KRVD--PFVQKIERGDFFVPEFENLGMQPLFAKNI--SYKYNNNTANSRIKNLGAFGWQP 479
Query 259 RYYNWKTKIDRIHGAFTTTLKDWVAPIDDSFLYSLFGDSLSTYKGVTWPFFKVNPNTLDD 318
RY +KT +D HG F P+ + G+S+S + T FK+NP LDD
Sbjct 480 RYSEYKTALDINHGQFVHQ-----EPLSYWTVARARGESMSNFNIST---FKINPKWLDD 531
Query 319 LFAVKVDSTWETDQLLVNCNVGCYVTRPLSADGVP 353
+FAV + T TDQ+ C +S DG+P
Sbjct 532 VFAVNYNGTELTDQVFGGCYFNIVKVSDMSIDGMP 566
>gi|494308783|ref|WP_007173938.1| hypothetical protein [Prevotella bergensis]
gi|270333035|gb|EFA43821.1| putative capsid protein (F protein) [Prevotella bergensis DSM
17361]
Length=553
Score = 119 bits (299), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 84/275 (31%), Positives = 128/275 (47%), Gaps = 31/275 (11%)
Query 91 FSVLALRQAEALQKWKEITQSVDTNYRDQIKAHFGVSVPQSEAHMAKYIGGVARNLDISE 150
FSV +LR A A+ K +T ++DQ++AH+GV +P S Y+GG ++ +S+
Sbjct 296 FSVSSLRAAFAVDKLLSVTMRAGKTFQDQMRAHYGVEIPDSRDGRVNYLGGFDSDMQVSD 355
Query 151 VVN-------NFLPGPGNDSSQAYIYGKGVGSGQGSMTFNTGSGYYILMCIYHAVPLLDY 203
V + P G + GKG GSG+G + F+ + +LMCIY VP + Y
Sbjct 356 VTQTSGTTATEYKPEAG---YLGRVAGKGTGSGRGRIVFDAKE-HGVLMCIYSLVPQIQY 411
Query 204 SISGPDGQNLVTSVEDLPIPEFDNIGMESVPAVELMNSSLYSNVNSAD---KILGYSPRY 260
+ D D PEF+N+GM+ +NSS S+ + D +LGY PRY
Sbjct 412 DCTRLDPMVDKLDRFDYFTPEFENLGMQP------LNSSYISSFCTTDPKNPVLGYQPRY 465
Query 261 YNWKTKIDRIHGAFTTT--LKDWVAPIDDSFLYSLFGDSLSTYKGVTWPFFKVNPNTLDD 318
+KT +D HG F + L W S F +T+ + FK++P L+
Sbjct 466 SEYKTALDVNHGQFAQSDALSSWSV--------SRF-RRWTTFPQLEIADFKIDPGCLNS 516
Query 319 LFAVKVDSTWETDQLLVNCNVGCYVTRPLSADGVP 353
+F V + T D + CN +S DG+P
Sbjct 517 IFPVDYNGTEANDCVYGGCNFNIVKVSDMSVDGMP 551
Lambda K H a alpha
0.315 0.134 0.398 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 2103429244710