bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-41_CDS_annotation_glimmer3.pl_2_1
Length=190
Score E
Sequences producing significant alignments: (Bits) Value
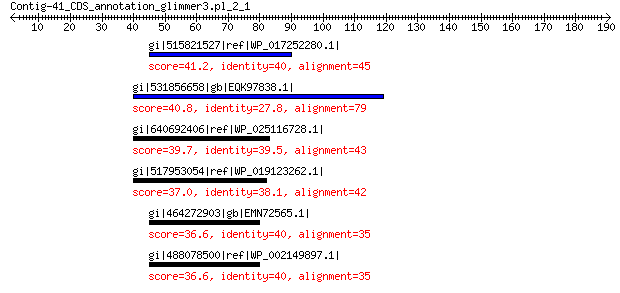
gi|515821527|ref|WP_017252280.1| pyridoxal phosphate-dependent a... 41.2 0.28
gi|531856658|gb|EQK97838.1| L-galactonate dehydratase 40.8 0.35
gi|640692406|ref|WP_025116728.1| aminotransferase DegT 39.7 0.76
gi|517953054|ref|WP_019123262.1| hypothetical protein 37.0 6.9
gi|464272903|gb|EMN72565.1| aminotransferase, LLPSF_NHT_00031 fa... 36.6 7.6
gi|488078500|ref|WP_002149897.1| aminotransferase, LLPSF_NHT_000... 36.6 7.9
>gi|515821527|ref|WP_017252280.1| pyridoxal phosphate-dependent aminotransferase [Brevibacillus
brevis]
Length=393
Score = 41.2 bits (95), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 18/45 (40%), Positives = 27/45 (60%), Gaps = 0/45 (0%)
Query 45 NDTVRYEYRVRSRNAIVSCNQLISIPEFIRSTRSFTDFITFATSR 89
+D V + YR+ + NA + C QL +P+FIR+ RS D A S+
Sbjct 258 HDQVGFNYRLPNINAALGCAQLERLPQFIRNKRSLADMYQAALSK 302
>gi|531856658|gb|EQK97838.1| L-galactonate dehydratase [Ophiocordyceps sinensis CO18]
Length=423
Score = 40.8 bits (94), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 22/79 (28%), Positives = 34/79 (43%), Gaps = 3/79 (4%)
Query 40 RTGVHNDTVRYEYRVRSRNAIVSCNQLISIPEFIRSTRSFTDFITFATSRPTQTVSLHGH 99
R + D + Y+ R +V NQ+ S+PE + R +DF + PT + GH
Sbjct 205 RLSIARDIIGYD---RGNVLMVDANQVWSVPEAVEHIRHLSDFKPWFIEEPTSPDDIVGH 261
Query 100 STTLTPVIPLEFRSACGLL 118
T + PL A G +
Sbjct 262 KTIREALKPLGIGVATGEM 280
>gi|640692406|ref|WP_025116728.1| aminotransferase DegT [Lysinibacillus fusiformis]
Length=384
Score = 39.7 bits (91), Expect = 0.76, Method: Compositional matrix adjust.
Identities = 17/43 (40%), Positives = 25/43 (58%), Gaps = 0/43 (0%)
Query 40 RTGVHNDTVRYEYRVRSRNAIVSCNQLISIPEFIRSTRSFTDF 82
R +D V + YR+ + NA + C QL +PEFI+ R+ T F
Sbjct 249 RWEYEHDEVAFNYRLPNINAALGCAQLEKMPEFIKRKRALTAF 291
>gi|517953054|ref|WP_019123262.1| hypothetical protein [Brevibacillus massiliensis]
Length=395
Score = 37.0 bits (84), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 16/42 (38%), Positives = 23/42 (55%), Gaps = 0/42 (0%)
Query 40 RTGVHNDTVRYEYRVRSRNAIVSCNQLISIPEFIRSTRSFTD 81
R ++D V Y YR+ + NA + C QL +P FI R+ D
Sbjct 253 RWAFNHDRVGYNYRLPNLNAALGCAQLEQLPGFIAKKRNLAD 294
>gi|464272903|gb|EMN72565.1| aminotransferase, LLPSF_NHT_00031 family [Leptospira interrogans
serovar Bataviae str. UI 08561]
Length=323
Score = 36.6 bits (83), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 14/35 (40%), Positives = 22/35 (63%), Gaps = 0/35 (0%)
Query 45 NDTVRYEYRVRSRNAIVSCNQLISIPEFIRSTRSF 79
+D + Y YR+ + NA + C QL +PEF++ SF
Sbjct 258 HDEIGYNYRMPNINAALGCAQLEQMPEFLKKNESF 292
>gi|488078500|ref|WP_002149897.1| aminotransferase, LLPSF_NHT_00031 family, partial [Leptospira
interrogans]
gi|464350066|gb|EMO29189.1| aminotransferase, LLPSF_NHT_00031 family, partial [Leptospira
interrogans serovar Bataviae str. HAI135]
Length=328
Score = 36.6 bits (83), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 14/35 (40%), Positives = 22/35 (63%), Gaps = 0/35 (0%)
Query 45 NDTVRYEYRVRSRNAIVSCNQLISIPEFIRSTRSF 79
+D + Y YR+ + NA + C QL +PEF++ SF
Sbjct 258 HDEIGYNYRMPNINAALGCAQLEQMPEFLKKNESF 292
Lambda K H a alpha
0.322 0.134 0.404 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 548287872192