bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-42_CDS_annotation_glimmer3.pl_2_1
Length=79
Score E
Sequences producing significant alignments: (Bits) Value
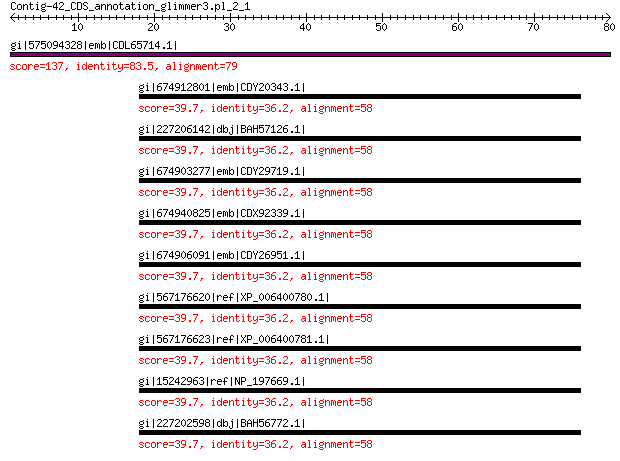
gi|575094328|emb|CDL65714.1| unnamed protein product 137 4e-40
gi|674912801|emb|CDY20343.1| BnaC01g23970D 39.7 0.086
gi|227206142|dbj|BAH57126.1| AT5G22770 39.7 0.13
gi|674903277|emb|CDY29719.1| BnaAnng03410D 39.7 0.13
gi|674940825|emb|CDX92339.1| BnaA10g13780D 39.7 0.13
gi|674906091|emb|CDY26951.1| BnaC09g36320D 39.7 0.13
gi|567176620|ref|XP_006400780.1| hypothetical protein EUTSA_v100... 39.7 0.13
gi|567176623|ref|XP_006400781.1| hypothetical protein EUTSA_v100... 39.7 0.13
gi|15242963|ref|NP_197669.1| alpha-adaptin 39.7 0.13
gi|227202598|dbj|BAH56772.1| AT5G22770 39.7 0.13
>gi|575094328|emb|CDL65714.1| unnamed protein product [uncultured bacterium]
Length=79
Score = 137 bits (346), Expect = 4e-40, Method: Compositional matrix adjust.
Identities = 66/79 (84%), Positives = 71/79 (90%), Gaps = 0/79 (0%)
Query 1 MKRKFYGTVKIQRFFGIKAGRSVWTDAYVHTRHYDTEFEAREALVFLVDSVLPATRVKIL 60
M+RKFYGTVKIQRFFGIK GRSVWT++YV TR+Y +E EAREAL FLVDSVLPATRVKIL
Sbjct 1 MQRKFYGTVKIQRFFGIKTGRSVWTESYVRTRYYFSECEAREALAFLVDSVLPATRVKIL 60
Query 61 GYIVESCFLPSSDDFDLPF 79
GY VESC LPS DDFDLPF
Sbjct 61 GYYVESCILPSPDDFDLPF 79
>gi|674912801|emb|CDY20343.1| BnaC01g23970D [Brassica napus]
Length=312
Score = 39.7 bits (91), Expect = 0.086, Method: Composition-based stats.
Identities = 21/58 (36%), Positives = 28/58 (48%), Gaps = 3/58 (5%)
Query 18 KAGRSVWTDAYVHTRHYDTEFEAREALVFLVDSVLPATRVKILGYIVESCFLPSSDDF 75
K + VW Y+H YD +F EA+ + P +V GYIV SC L + DF
Sbjct 48 KKKKYVWKMLYIHMLGYDVDFGHMEAVSLISAPKYPEKQV---GYIVTSCLLNENHDF 102
>gi|227206142|dbj|BAH57126.1| AT5G22770 [Arabidopsis thaliana]
Length=932
Score = 39.7 bits (91), Expect = 0.13, Method: Composition-based stats.
Identities = 21/58 (36%), Positives = 28/58 (48%), Gaps = 3/58 (5%)
Query 18 KAGRSVWTDAYVHTRHYDTEFEAREALVFLVDSVLPATRVKILGYIVESCFLPSSDDF 75
K + VW Y+H YD +F EA+ + P +V GYIV SC L + DF
Sbjct 48 KKKKYVWKMLYIHMLGYDVDFGHMEAVSLISAPKYPEKQV---GYIVTSCLLNENHDF 102
>gi|674903277|emb|CDY29719.1| BnaAnng03410D [Brassica napus]
Length=981
Score = 39.7 bits (91), Expect = 0.13, Method: Composition-based stats.
Identities = 21/58 (36%), Positives = 28/58 (48%), Gaps = 3/58 (5%)
Query 18 KAGRSVWTDAYVHTRHYDTEFEAREALVFLVDSVLPATRVKILGYIVESCFLPSSDDF 75
K + VW Y+H YD +F EA+ + P +V GYIV SC L + DF
Sbjct 48 KKKKYVWKMLYIHMLGYDVDFGHMEAVSLISAPKYPEKQV---GYIVTSCLLNENHDF 102
>gi|674940825|emb|CDX92339.1| BnaA10g13780D [Brassica napus]
Length=1007
Score = 39.7 bits (91), Expect = 0.13, Method: Composition-based stats.
Identities = 21/58 (36%), Positives = 28/58 (48%), Gaps = 3/58 (5%)
Query 18 KAGRSVWTDAYVHTRHYDTEFEAREALVFLVDSVLPATRVKILGYIVESCFLPSSDDF 75
K + VW Y+H YD +F EA+ + P +V GYIV SC L + DF
Sbjct 48 KKKKYVWKMLYIHMLGYDVDFGHMEAVSLISAPKYPEKQV---GYIVTSCLLNENHDF 102
>gi|674906091|emb|CDY26951.1| BnaC09g36320D [Brassica napus]
Length=1008
Score = 39.7 bits (91), Expect = 0.13, Method: Composition-based stats.
Identities = 21/58 (36%), Positives = 28/58 (48%), Gaps = 3/58 (5%)
Query 18 KAGRSVWTDAYVHTRHYDTEFEAREALVFLVDSVLPATRVKILGYIVESCFLPSSDDF 75
K + VW Y+H YD +F EA+ + P +V GYIV SC L + DF
Sbjct 48 KKKKYVWKMLYIHMLGYDVDFGHMEAVSLISAPKYPEKQV---GYIVTSCLLNENHDF 102
>gi|567176620|ref|XP_006400780.1| hypothetical protein EUTSA_v10012556mg [Eutrema salsugineum]
gi|557101870|gb|ESQ42233.1| hypothetical protein EUTSA_v10012556mg [Eutrema salsugineum]
Length=1011
Score = 39.7 bits (91), Expect = 0.13, Method: Composition-based stats.
Identities = 21/58 (36%), Positives = 28/58 (48%), Gaps = 3/58 (5%)
Query 18 KAGRSVWTDAYVHTRHYDTEFEAREALVFLVDSVLPATRVKILGYIVESCFLPSSDDF 75
K + VW Y+H YD +F EA+ + P +V GYIV SC L + DF
Sbjct 48 KKKKYVWKMLYIHMLGYDVDFGHMEAVSLISAPKYPEKQV---GYIVTSCLLNENHDF 102
>gi|567176623|ref|XP_006400781.1| hypothetical protein EUTSA_v10012556mg [Eutrema salsugineum]
gi|557101871|gb|ESQ42234.1| hypothetical protein EUTSA_v10012556mg [Eutrema salsugineum]
Length=1012
Score = 39.7 bits (91), Expect = 0.13, Method: Composition-based stats.
Identities = 21/58 (36%), Positives = 28/58 (48%), Gaps = 3/58 (5%)
Query 18 KAGRSVWTDAYVHTRHYDTEFEAREALVFLVDSVLPATRVKILGYIVESCFLPSSDDF 75
K + VW Y+H YD +F EA+ + P +V GYIV SC L + DF
Sbjct 48 KKKKYVWKMLYIHMLGYDVDFGHMEAVSLISAPKYPEKQV---GYIVTSCLLNENHDF 102
>gi|15242963|ref|NP_197669.1| alpha-adaptin [Arabidopsis thaliana]
gi|30688634|ref|NP_851057.1| alpha-adaptin [Arabidopsis thaliana]
gi|30688640|ref|NP_851058.1| alpha-adaptin [Arabidopsis thaliana]
gi|75157392|sp|Q8LPL6.1|AP2A1_ARATH RecName: Full=AP-2 complex subunit alpha-1; AltName: Full=Adaptor
protein complex AP-2 subunit alpha-1; AltName: Full=Adaptor-related
protein complex 2 subunit alpha-1; AltName: Full=Alpha-adaptin
1; AltName: Full=Clathrin assembly protein complex
2 alpha large chain 1; Short=At-a-Ad; Short=At-alpha-Ad
[Arabidopsis thaliana]
gi|20466205|gb|AAM20420.1| alpha-adaptin [Arabidopsis thaliana]
gi|332005689|gb|AED93072.1| alpha-adaptin [Arabidopsis thaliana]
gi|332005690|gb|AED93073.1| alpha-adaptin [Arabidopsis thaliana]
gi|332005691|gb|AED93074.1| alpha-adaptin [Arabidopsis thaliana]
Length=1012
Score = 39.7 bits (91), Expect = 0.13, Method: Composition-based stats.
Identities = 21/58 (36%), Positives = 28/58 (48%), Gaps = 3/58 (5%)
Query 18 KAGRSVWTDAYVHTRHYDTEFEAREALVFLVDSVLPATRVKILGYIVESCFLPSSDDF 75
K + VW Y+H YD +F EA+ + P +V GYIV SC L + DF
Sbjct 48 KKKKYVWKMLYIHMLGYDVDFGHMEAVSLISAPKYPEKQV---GYIVTSCLLNENHDF 102
>gi|227202598|dbj|BAH56772.1| AT5G22770 [Arabidopsis thaliana]
Length=1013
Score = 39.7 bits (91), Expect = 0.13, Method: Composition-based stats.
Identities = 21/58 (36%), Positives = 28/58 (48%), Gaps = 3/58 (5%)
Query 18 KAGRSVWTDAYVHTRHYDTEFEAREALVFLVDSVLPATRVKILGYIVESCFLPSSDDF 75
K + VW Y+H YD +F EA+ + P +V GYIV SC L + DF
Sbjct 48 KKKKYVWKMLYIHMLGYDVDFGHMEAVSLISAPKYPEKQV---GYIVTSCLLNENHDF 102
Lambda K H a alpha
0.328 0.143 0.441 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 438269475921