bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-45_CDS_annotation_glimmer3.pl_2_1
Length=188
Score E
Sequences producing significant alignments: (Bits) Value
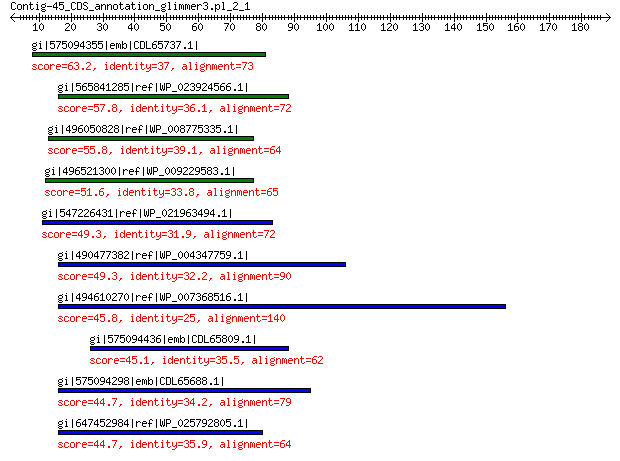
gi|575094355|emb|CDL65737.1| unnamed protein product 63.2 1e-08
gi|565841285|ref|WP_023924566.1| hypothetical protein 57.8 6e-07
gi|496050828|ref|WP_008775335.1| hypothetical protein 55.8 3e-06
gi|496521300|ref|WP_009229583.1| hypothetical protein 51.6 9e-05
gi|547226431|ref|WP_021963494.1| predicted protein 49.3 5e-04
gi|490477382|ref|WP_004347759.1| hypothetical protein 49.3 5e-04
gi|494610270|ref|WP_007368516.1| hypothetical protein 45.8 0.006
gi|575094436|emb|CDL65809.1| unnamed protein product 45.1 0.012
gi|575094298|emb|CDL65688.1| unnamed protein product 44.7 0.019
gi|647452984|ref|WP_025792805.1| hypothetical protein 44.7 0.020
>gi|575094355|emb|CDL65737.1| unnamed protein product [uncultured bacterium]
Length=517
Score = 63.2 bits (152), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 27/73 (37%), Positives = 44/73 (60%), Gaps = 0/73 (0%)
Query 8 LTKYLFTECLRPQRIVNPYSHDVIFAPCGRCKSCIMNKSNFATAYAMNMATHFKYCYFVT 67
L + F CL P+R+ NPY +D + PCG+C++C +K++ A+ K+C F T
Sbjct 3 LKDFPFIRCLEPKRVFNPYLNDWLLVPCGKCRACQCSKASRYKLQIQLEASQHKFCIFGT 62
Query 68 LTYKDIFLPYLSV 80
LTY + ++P LS+
Sbjct 63 LTYANTYIPRLSL 75
>gi|565841285|ref|WP_023924566.1| hypothetical protein [Prevotella nigrescens]
gi|564729906|gb|ETD29850.1| hypothetical protein HMPREF1173_00032 [Prevotella nigrescens
CC14M]
Length=484
Score = 57.8 bits (138), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 26/72 (36%), Positives = 38/72 (53%), Gaps = 0/72 (0%)
Query 16 CLRPQRIVNPYSHDVIFAPCGRCKSCIMNKSNFATAYAMNMATHFKYCYFVTLTYKDIFL 75
C P+RI+NPY+H+ ++ C RCK C+ K++ + N Y FVTLTY + L
Sbjct 9 CEHPKRIINPYTHERVWVACRRCKCCLNKKTSAWSGRVANECKLHAYSAFVTLTYDNEHL 68
Query 76 PYLSVEVVRRSG 87
P E + G
Sbjct 69 PLYQPECMNERG 80
>gi|496050828|ref|WP_008775335.1| hypothetical protein [Bacteroides sp. 2_2_4]
gi|229448892|gb|EEO54683.1| hypothetical protein BSCG_01608 [Bacteroides sp. 2_2_4]
Length=497
Score = 55.8 bits (133), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 25/65 (38%), Positives = 43/65 (66%), Gaps = 2/65 (3%)
Query 13 FTECLRPQRIVNPYSHDVIFAPCGRCKSCIMNKSNFATAYAMNMATH-FKYCYFVTLTYK 71
F +CL P+RI+NPY+ + + PCG C++C + K N A+ ++ ++ K+ F+TLTY
Sbjct 6 FCKCLHPKRIMNPYTKESMVVPCGHCQACTLAK-NSRYAFQCDLESYTAKHTLFITLTYA 64
Query 72 DIFLP 76
+ F+P
Sbjct 65 NRFIP 69
>gi|496521300|ref|WP_009229583.1| hypothetical protein [Prevotella sp. oral taxon 317]
gi|288330571|gb|EFC69155.1| hypothetical protein HMPREF0670_00478 [Prevotella sp. oral taxon
317 str. F0108]
Length=569
Score = 51.6 bits (122), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 22/65 (34%), Positives = 35/65 (54%), Gaps = 0/65 (0%)
Query 12 LFTECLRPQRIVNPYSHDVIFAPCGRCKSCIMNKSNFATAYAMNMATHFKYCYFVTLTYK 71
+F CL P + N ++ D +F PCGRC++C+ ++ + N KY TLTY
Sbjct 7 IFGNCLCPVHVHNRWTRDEMFVPCGRCEACVNAAASKQSKRVRNEIMQHKYSVMFTLTYN 66
Query 72 DIFLP 76
+ F+P
Sbjct 67 NEFIP 71
>gi|547226431|ref|WP_021963494.1| predicted protein [Prevotella sp. CAG:1185]
gi|524103383|emb|CCY83995.1| predicted protein [Prevotella sp. CAG:1185]
Length=498
Score = 49.3 bits (116), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 23/72 (32%), Positives = 38/72 (53%), Gaps = 0/72 (0%)
Query 11 YLFTECLRPQRIVNPYSHDVIFAPCGRCKSCIMNKSNFATAYAMNMATHFKYCYFVTLTY 70
Y +C P+ + N Y+ +VI CG CK+C+ +++ + KYC F TLTY
Sbjct 6 YPLVKCYHPRHVQNKYTGEVIQVGCGVCKACLKRRADKMSFLCAIEEQSHKYCMFATLTY 65
Query 71 KDIFLPYLSVEV 82
+ ++P + EV
Sbjct 66 SNDYVPRMYPEV 77
>gi|490477382|ref|WP_004347759.1| hypothetical protein [Prevotella buccalis]
gi|281300711|gb|EFA93042.1| hypothetical protein HMPREF0650_1078 [Prevotella buccalis ATCC
35310]
Length=582
Score = 49.3 bits (116), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 29/95 (31%), Positives = 45/95 (47%), Gaps = 5/95 (5%)
Query 16 CLRPQRIVNPYSHDVIFAPCGRCKSCIMNKSNFATAYAMNMATHFKYCYFVTLTYKDIFL 75
CL P+++ NP H ++ C +C +C+ K+ + A KY F TLTY + L
Sbjct 13 CLNPRKVYNPSLHGWMYCSCDKCTACLNQKATTLSNRARAEIEQHKYSVFFTLTYDNEHL 72
Query 76 PYLSV-----EVVRRSGNRYLFDENFETMVSTSDP 105
P V EV++ L D++ M+S S P
Sbjct 73 PKYEVFQDSNEVIQYRPIGRLVDDSSSDMLSNSCP 107
>gi|494610270|ref|WP_007368516.1| hypothetical protein [Prevotella multiformis]
gi|324988542|gb|EGC20505.1| hypothetical protein HMPREF9141_0984 [Prevotella multiformis
DSM 16608]
Length=479
Score = 45.8 bits (107), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 35/146 (24%), Positives = 61/146 (42%), Gaps = 9/146 (6%)
Query 16 CLRPQRIVNPYSHDVIFAPCGRCKSCIMNKSNFATAYAM---NMATHFKYCYFVTLTYKD 72
CL P+RI N Y + ++ PC +C C + ++A+ ++ N ++ FVTLTY +
Sbjct 10 CLSPKRIYNKYIDETLYVPCRKCFRC---RDSYASDWSRRIENECREHRFSLFVTLTYDN 66
Query 73 IFLPYLSVEVVRRSGNRYLFDENFE---TMVSTSDPRLLTPEYYHDRDLSLDPAQNEVEQ 129
+P V+ + F +S S R L P+ D P + +V+
Sbjct 67 EHIPLFQPLVMDDGSHPVWFSNRLSESGKFLSDSVCRSLPPQKMEDEVCFAYPCKKDVQD 126
Query 130 VFDIGFQSIPRDVSVKSKGSFRFRSF 155
F ++ ++ FR R F
Sbjct 127 WFKRLRSAVDYQLNKNKSNEFRIRYF 152
>gi|575094436|emb|CDL65809.1| unnamed protein product [uncultured bacterium]
Length=340
Score = 45.1 bits (105), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 22/63 (35%), Positives = 34/63 (54%), Gaps = 5/63 (8%)
Query 26 YSHDVIFAPCGRCKSC-IMNKSNFATAYAMNMATHFKYCYFVTLTYKDIFLPYLSVEVVR 84
+ DV+ PCG+C C I K ++AT + + +FVTLTY+D +P L +R
Sbjct 42 HRQDVMLIPCGKCIGCRIRAKQDWATRLELEARAYKGRAWFVTLTYRDDTIPLL----IR 97
Query 85 RSG 87
+G
Sbjct 98 NTG 100
>gi|575094298|emb|CDL65688.1| unnamed protein product [uncultured bacterium]
Length=478
Score = 44.7 bits (104), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 27/82 (33%), Positives = 44/82 (54%), Gaps = 4/82 (5%)
Query 16 CLRPQRIVNPYSHDVIFAPCGRCKSCIMNKSNFATAYAM-NMATHFKYCYFVTLTYKDIF 74
C+ + I N Y+ ++ CG+C +C+ K+N A+AY + N + C+FVTL Y +
Sbjct 2 CINKREIRNKYTGQKLYVSCGKCPACLQEKAN-ASAYKIRNNQSSELSCFFVTLNYDNNH 60
Query 75 LPYLSVEVV--RRSGNRYLFDE 94
+P + V S + Y FDE
Sbjct 61 IPVIFKHDVYNYNSSDVYHFDE 82
>gi|647452984|ref|WP_025792805.1| hypothetical protein [Prevotella histicola]
Length=480
Score = 44.7 bits (104), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 23/67 (34%), Positives = 39/67 (58%), Gaps = 6/67 (9%)
Query 16 CLRPQRIVNPYSHDVIFAPCGRCKSCIMNKSNFATAYAMNM---ATHFKYCYFVTLTYKD 72
CLRP RI N Y + ++ C +C C +S++A+++A + + +Y F+TLTY +
Sbjct 12 CLRPHRIYNRYIGEFLYTNCRKCVRC---RSSYASSWANRIDSECSFHRYSLFLTLTYDN 68
Query 73 IFLPYLS 79
LPY +
Sbjct 69 DHLPYYA 75
Lambda K H a alpha
0.325 0.140 0.427 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 526786387008