bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-8_CDS_annotation_glimmer3.pl_2_7
Length=322
Score E
Sequences producing significant alignments: (Bits) Value
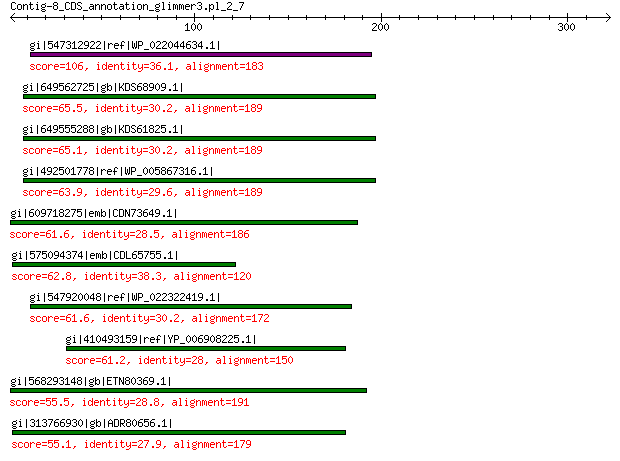
gi|547312922|ref|WP_022044634.1| putative replication initiation... 106 5e-23
gi|649562725|gb|KDS68909.1| hypothetical protein M096_3339 65.5 4e-09
gi|649555288|gb|KDS61825.1| hypothetical protein M095_3808 65.1 9e-09
gi|492501778|ref|WP_005867316.1| hypothetical protein 63.9 2e-08
gi|609718275|emb|CDN73649.1| conserved hypothetical protein 61.6 1e-07
gi|575094374|emb|CDL65755.1| unnamed protein product 62.8 1e-07
gi|547920048|ref|WP_022322419.1| putative replication protein 61.6 1e-07
gi|410493159|ref|YP_006908225.1| replication-associated protein 61.2 2e-07
gi|568293148|gb|ETN80369.1| hypothetical protein NECAME_18023 55.5 2e-05
gi|313766930|gb|ADR80656.1| putative replication initiation protein 55.1 3e-05
>gi|547312922|ref|WP_022044634.1| putative replication initiation protein [Alistipes finegoldii
CAG:68]
gi|524208442|emb|CCZ76638.1| putative replication initiation protein [Alistipes finegoldii
CAG:68]
Length=320
Score = 106 bits (265), Expect = 5e-23, Method: Compositional matrix adjust.
Identities = 66/196 (34%), Positives = 99/196 (51%), Gaps = 29/196 (15%)
Query 12 FTTLTISPEGIKEIEEKTGLKWEENPNEIATKGLRLFLERVRKETGKSIKHWCVTELGEK 71
F TLT + + +++ + T K +RLFL+R RK GK I+HW V E G
Sbjct 76 FVTLTFNDDSLEKFSKDT------------NKAVRLFLDRFRKVYGKQIRHWFVCEFGTL 123
Query 72 KDRIHLHGIFFGQKSA-------------ELIRKHWKYGFVFIGGYCNSRSVNYITKYML 118
R H HGI F A L+ WKYGFVF+ GY + + +YITKY+
Sbjct 124 HGRPHYHGILFNVPQALIDGYDSDMPGHHPLLASCWKYGFVFV-GYVSDETCSYITKYVT 182
Query 119 KVDIKHPEFKQIVLASAGIGAGYMDRLDYLWQKQNYKNINVATYTFRNGTKMAMPKYYKN 178
K I + + V++S GIG+ Y++ + K N + NG + AMP+YY N
Sbjct 183 K-SINGDKVRPRVISSFGIGSNYLNTEESSLHKLG--NQRYQPFMVLNGFQQAMPRYYYN 239
Query 179 KIFTEKEREKMWINNL 194
KIF++ +++ M ++ L
Sbjct 240 KIFSDVDKQNMVVDRL 255
>gi|649562725|gb|KDS68909.1| hypothetical protein M096_3339 [Parabacteroides distasonis str.
3999B T(B) 6]
Length=250
Score = 65.5 bits (158), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 57/211 (27%), Positives = 104/211 (49%), Gaps = 29/211 (14%)
Query 8 SFGYFTTLTISPEGIKEIEEKTGLKWEENPNEIATKGLRLFLERVRKETGK-SIKHWCVT 66
F F TLT E I L ++ ++ + ++LF++R+RK+ + ++++ +
Sbjct 9 PFSLFVTLTYDDEHIPTAMIGEDL-FKTTVGVVSKRDIQLFMKRLRKKYAQYRLRYFLTS 67
Query 67 ELGEKKDRIHLHGIFFG-----QKSAELIRKHWKYGFVFIGGYCNSRSVNYITKYM---- 117
E G + R H H I FG + +L+ + WK GFV ++ ++Y+TKYM
Sbjct 68 EYGSQGGRPHYHMILFGFPFTGKHGGDLLAECWKNGFV-QAHPLTTKEISYVTKYMYEKS 126
Query 118 LKVDIKH--PEFKQIVLASAGIGAGYMDRLDYLWQKQ--NYKNINVATYTFR-NGTKMAM 172
+ DI E++ +L S G GY + ++Q ++ ++ Y NG +MAM
Sbjct 127 MIPDILKGVKEYQPFMLCSKMPGIGY-----HFLREQILDFYRLHPRDYVRAFNGMRMAM 181
Query 173 PKYYKNKIFTE--KE-----REKMWINNLNR 196
P+YY +K++ + KE RE +IN + +
Sbjct 182 PRYYADKLYDDDMKEYLKELREAFFINQMQQ 212
>gi|649555288|gb|KDS61825.1| hypothetical protein M095_3808 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649560564|gb|KDS66872.1| hypothetical protein M095_2449 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649561011|gb|KDS67298.1| hypothetical protein M095_2409 [Parabacteroides distasonis str.
3999B T(B) 4]
Length=284
Score = 65.1 bits (157), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 57/211 (27%), Positives = 104/211 (49%), Gaps = 29/211 (14%)
Query 8 SFGYFTTLTISPEGIKEIEEKTGLKWEENPNEIATKGLRLFLERVRKETGK-SIKHWCVT 66
F F TLT E I L ++ ++ + ++LF++R+RK+ + ++++ +
Sbjct 43 PFSLFVTLTYDDEHIPTAMIGEDL-FKTTVGVVSKRDIQLFMKRLRKKYAQYRLRYFLTS 101
Query 67 ELGEKKDRIHLHGIFFG-----QKSAELIRKHWKYGFVFIGGYCNSRSVNYITKYM---- 117
E G + R H H I FG + +L+ + WK GFV ++ ++Y+TKYM
Sbjct 102 EYGSQGGRPHYHMILFGFPFTGKHGGDLLAECWKNGFV-QAHPLTTKEISYVTKYMYEKS 160
Query 118 LKVDIKH--PEFKQIVLASAGIGAGYMDRLDYLWQKQ--NYKNINVATYTFR-NGTKMAM 172
+ DI E++ +L S G GY + ++Q ++ ++ Y NG +MAM
Sbjct 161 MIPDILKGVKEYQPFMLCSKMPGIGY-----HFLREQILDFYRLHPRDYVRAFNGMRMAM 215
Query 173 PKYYKNKIFTE--KE-----REKMWINNLNR 196
P+YY +K++ + KE RE +IN + +
Sbjct 216 PRYYADKLYDDDMKEYLKELREAFFINQMQQ 246
>gi|492501778|ref|WP_005867316.1| hypothetical protein [Parabacteroides distasonis]
gi|409230407|gb|EKN23271.1| hypothetical protein HMPREF1059_03256 [Parabacteroides distasonis
CL09T03C24]
Length=284
Score = 63.9 bits (154), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 56/213 (26%), Positives = 104/213 (49%), Gaps = 33/213 (15%)
Query 8 SFGYFTTLTISPEGIKEIEEKTGLKWEENPNEIATKGLRLFLERVRKETGK-SIKHWCVT 66
F F TLT E + L ++ ++ + ++LF++R+RK+ + ++++ +
Sbjct 43 PFSLFVTLTYDDEHMPTAMIGEDL-FKSTVGVVSKRDIQLFMKRLRKKYDQYRLRYFLTS 101
Query 67 ELGEKKDRIHLHGIFFG-----QKSAELIRKHWKYGFVFIGGYCNSRSVNYITKYM---- 117
E G + R H H I FG + +L+ + WK GFV ++ + Y+TKYM
Sbjct 102 EYGSQGGRPHYHMILFGFPFTGKHGGDLLAECWKNGFV-QAHPLTTKEIAYVTKYMYEKS 160
Query 118 ----LKVDIKHPEFKQIVLASAGIGAGYMDRLDYLWQKQ--NYKNINVATYTFR-NGTKM 170
+ D+K E++ +L S G GY + ++Q ++ ++ Y NG +M
Sbjct 161 MVPDILKDVK--EYQPFMLCSRIPGIGY-----HFLREQILDFYRLHPRDYVRAFNGMRM 213
Query 171 AMPKYYKNKIFTE--KE-----REKMWINNLNR 196
AMP+YY +K++ + KE RE +IN + +
Sbjct 214 AMPRYYADKLYDDDMKEYLKELREAFFINQMQQ 246
>gi|609718275|emb|CDN73649.1| conserved hypothetical protein [Elizabethkingia anophelis]
Length=265
Score = 61.6 bits (148), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 53/195 (27%), Positives = 89/195 (46%), Gaps = 23/195 (12%)
Query 1 LEEELH-SSFGYFTTLTISPEGIKEIEEKTGLKWEENP-NEIATKGLRLFLERVRKETGK 58
L EEL S +F TLT S L + +N + + +LF++R RK
Sbjct 43 LTEELKVSKSAHFVTLTYS---------DVYLPYSDNGLISLDYRDFQLFMKRARKLQKS 93
Query 59 SIKHWCVTELGEKKDRIHLHGIFFGQKSAELIRKHWKYGFVFIGGYCNSRSVNYITKYML 118
IK++ V E G + R H H I FG ++ + W+ G V G ++S+ Y KY
Sbjct 94 KIKYFLVGEYGAQTYRPHYHAIVFGVENIDAFLGEWRMGNVH-AGTVTAKSIYYTLKYCT 152
Query 119 K-------VDIKHPEFKQIVLASAGIGAGYMDRLDYLWQKQNYKNINVATYTFRNGTKMA 171
K D + L S G+G ++ + YK+ +++ GT +A
Sbjct 153 KSITEGPDKDPDDDRKPEKALMSKGLGLSHLTE----SMIKYYKDDVSRSFSLLGGTTIA 208
Query 172 MPKYYKNKIFTEKER 186
+P+YY++K+F++ E+
Sbjct 209 LPRYYRDKVFSDIEK 223
>gi|575094374|emb|CDL65755.1| unnamed protein product [uncultured bacterium]
Length=487
Score = 62.8 bits (151), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 46/134 (34%), Positives = 67/134 (50%), Gaps = 18/134 (13%)
Query 2 EEELHSSFGYFTTLTISPEGIKEIEEKTGLKWEENPNEIATK-GLRLFLERVRKETGK-- 58
EE ++S YF TLT+ P I + G + +P + K ++LFL+R+RK K
Sbjct 48 EELNNNSQSYFYTLTLDPRFI----DTYGTLPDGSPRYVFNKRHIQLFLKRLRKALSKYN 103
Query 59 -SIKHWCVTELGEKKDRIHLHGIFFGQKSAE------LIRKHWKYGFVFIGG----YCNS 107
S+K+ V ELGE R H H IF+ S ++R W GF+ G N+
Sbjct 104 ISLKYVIVGELGETTHRPHYHAIFYLSSSVNPFKFRIMVRNSWSLGFIKSGDNNGIILNN 163
Query 108 RSVNYITKYMLKVD 121
+V+Y+ KYM K D
Sbjct 164 DAVSYVIKYMHKTD 177
>gi|547920048|ref|WP_022322419.1| putative replication protein [Parabacteroides merdae CAG:48]
gi|524592960|emb|CDD13572.1| putative replication protein [Parabacteroides merdae CAG:48]
Length=278
Score = 61.6 bits (148), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 52/187 (28%), Positives = 93/187 (50%), Gaps = 22/187 (12%)
Query 12 FTTLTISPEGIKEIEEKTGLKWEENPNEIATKGLRLFLERVRKE-TGKSIKHWCVTELGE 70
F TLT E + IE ++ N ++ + ++LF++R+RK+ ++++ +E G
Sbjct 42 FVTLTYDDEHLP-IERIGSDLFQTNVAVVSKRDVQLFMKRLRKKYEDYKMRYFVTSEYGA 100
Query 71 KKDRIHLHGIFFG-----QKSAELIRKHWKYGFVFIGGYCNSRSVNYITKYMLKVDIKHP 125
K R H H I FG + + +L+ + W+ GFV + + Y+ KYM + + P
Sbjct 101 KNGRPHYHMILFGFPFTGKMAGDLLAECWQNGFV-QAHPLTIKEIAYVCKYMYEKSM-CP 158
Query 126 E-------FKQIVLAS--AGIGAGYMDRLDYLWQKQNYKNINVATYTFRNGTKMAMPKYY 176
E +K +L S GIG G+M + D + + + V + G KMAMP+YY
Sbjct 159 EILRDEKKYKPFMLCSRNPGIGFGFM-KADIIEFYRRHPRDYVRAWA---GHKMAMPRYY 214
Query 177 KNKIFTE 183
+K++ +
Sbjct 215 ADKLYDD 221
>gi|410493159|ref|YP_006908225.1| replication-associated protein [Dragonfly-associated microphage
1]
gi|406870779|gb|AFS65317.1| replication-associated protein [Dragonfly-associated microphage
1]
Length=270
Score = 61.2 bits (147), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 42/165 (25%), Positives = 76/165 (46%), Gaps = 16/165 (10%)
Query 31 LKWEENPNEIATKGLRLFLERVRKETGKSIKHWCVTELGEKKDRIHLHGIFFGQKSAE-- 88
L + ++P E+ + + FL+R R + ++++ V E G+ + R H H FG E
Sbjct 50 LTYRDDPGELVPEDMTNFLKRFRYYLQRKVRYFGVGEYGDLRGRPHFHMALFGVSPLEEG 109
Query 89 LIRKHWKYGFVFIGGYCNSRS---VNYITKYMLK-----VDIKHPEFKQIVLASAGIGAG 140
+I K W GFV +G + Y+TK M K +D ++PEF ++ GIGA
Sbjct 110 IIAKAWSIGFVHVGDLTKDSAQYLCGYVTKKMTKAEDERLDGRYPEFARMS-KDPGIGAS 168
Query 141 YMDRLDYLWQKQN-----YKNINVATYTFRNGTKMAMPKYYKNKI 180
+ L ++N +V + G ++ + +Y + K+
Sbjct 169 ALPVLREALAPDGDVTLMHRNGDVPSTMRTRGKELPLGRYLRGKL 213
>gi|568293148|gb|ETN80369.1| hypothetical protein NECAME_18023 [Necator americanus]
Length=345
Score = 55.5 bits (132), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 55/211 (26%), Positives = 90/211 (43%), Gaps = 41/211 (19%)
Query 1 LEEELHSSFGYFTTLT-------ISPEGIKEIEEKTGLKWEENPNEIATKGLRLFLERVR 53
L+EEL F TLT IS G ++ E P +++R+R
Sbjct 41 LQEELQHENASFVTLTYDTRFVPISKNGFMTLDRG------EFPR---------YMKRLR 85
Query 54 K-ETGKSIKHWCVTELGEKKDRIHLHGIFFGQKSAELIRKHWKYG----FVFIGGYCNSR 108
K G+ +K++ E G ++ R H H I FG L W + G +
Sbjct 86 KLVPGRKLKYYMCGEYGSQRFRPHYHAIIFGVPQDSLFADAWTLNGDSLGGVVVGTVTGK 145
Query 109 SVNYITKYMLKV--------DIKHPEFKQIVLASAGIGAGYMDRLDYLWQKQNYKNINVA 160
S+ Y KY+ K D + PEF L S G+G Y+ + K+ +I+
Sbjct 146 SIAYTMKYIDKSTWKQKHGRDDRVPEFS---LMSKGMGVSYLTPQMVEYHKE---DISRL 199
Query 161 TYTFRNGTKMAMPKYYKNKIFTEKEREKMWI 191
T G+++AMP+YY+ KI+++ + +K +
Sbjct 200 FCTREGGSRIAMPRYYRQKIYSDDDLKKQVV 230
>gi|313766930|gb|ADR80656.1| putative replication initiation protein [Uncultured Microviridae]
Length=402
Score = 55.1 bits (131), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 50/213 (23%), Positives = 86/213 (40%), Gaps = 46/213 (22%)
Query 2 EEELHSSFGYFTTLTISPEGIKEIEEKTGLKWEENPNEIATKGLRLFLERVRKETGKSIK 61
E ++H F TLTI+PE ++ P + K + F+ R+R++ GK IK
Sbjct 151 EAQMHDH-NCFITLTINPETLER---------RPRPWSLEKKEFQEFVHRLRRKIGKKIK 200
Query 62 HWCVTELGEKKDRIHLHGIFFGQK-----------------SAELIRKHWKYGFVFIGGY 104
++ E G++ R H H I FG S EL W +G+ IG
Sbjct 201 YFHCGEYGDENKRPHYHAIIFGYDFPDKQLWERKLGNELYISPEL-ENLWPHGYHRIGA- 258
Query 105 CNSRSVNYITKYMLKV----------------DIKHPEFKQIVLASAGIGAGYMDRLDYL 148
C S +Y+ +Y++K ++++ Q S G + +
Sbjct 259 CTYESAHYVARYVMKRAKGEGPPEQYINPETGEVEYDLDNQYATMSRGNKKQPQNGIGNQ 318
Query 149 WQ-KQNYKNINVATYTFRNGTKMAMPKYYKNKI 180
W K + + + Y +G KM +P+YY ++
Sbjct 319 WYWKYGWTDAHCHDYIVHDGIKMKVPRYYDKEL 351
Lambda K H a alpha
0.319 0.135 0.420 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1793877651450