bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-11_CDS_annotation_glimmer3.pl_2_1
Length=119
Score E
Sequences producing significant alignments: (Bits) Value
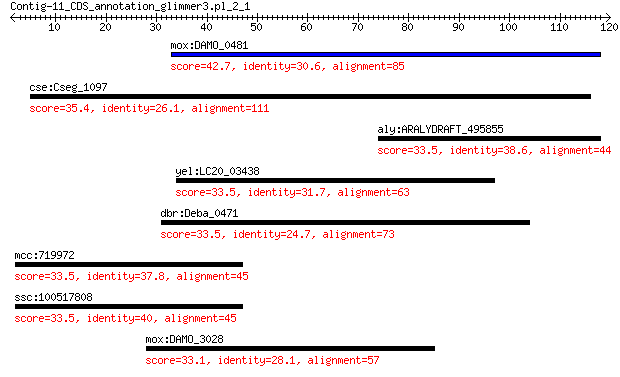
mox:DAMO_0481 hypothetical protein 42.7 0.004
cse:Cseg_1097 hypothetical protein 35.4 1.3
aly:ARALYDRAFT_495855 calmodulin-binding protein 33.5 7.4
yel:LC20_03438 peptidase (caspase-1 like protein) 33.5 7.6
dbr:Deba_0471 hypothetical protein 33.5 7.7
mcc:719972 niban-like protein 2-like 33.5 8.0
ssc:100517808 FAM129C; family with sequence similarity 129, me... 33.5 8.9
mox:DAMO_3028 hypothetical protein 33.1 9.7
> mox:DAMO_0481 hypothetical protein
Length=234
Score = 42.7 bits (99), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 26/85 (31%), Positives = 43/85 (51%), Gaps = 1/85 (1%)
Query 33 NIDTVHAGFRAGAAYQSWVAPLDGWNVLTSSGAWSYQSMKVRPQQLNSIFVPQVDSANCS 92
+I+ V A R GAA+ + PL VL S A +++ K+R Q + V + +
Sbjct 86 SIENVIAAMRRGAAFDYLLKPLQDLTVLEVSVARAFEIRKLRAQAREAFQVGAIRELAVT 145
Query 93 VAFDQLLCNVNFQVYAVQNLDRNGL 117
A D++L +N +V+ L RNG+
Sbjct 146 -ASDRILNPLNIISLSVERLTRNGM 169
> cse:Cseg_1097 hypothetical protein
Length=312
Score = 35.4 bits (80), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 29/111 (26%), Positives = 43/111 (39%), Gaps = 12/111 (11%)
Query 5 PSLNLQNNPGRNVSGALGYNLRYWQWKSNIDTVHAGFRAGAAYQSWVAPLDGWNVLTSSG 64
P + N R SG NL YW W S + V GA +++ + P +
Sbjct 65 PDPEMMNEFVRAQSGMTALNLLYWLWSSFVRAVL----CGAVFRAVLMP--------EAS 112
Query 65 AWSYQSMKVRPQQLNSIFVPQVDSANCSVAFDQLLCNVNFQVYAVQNLDRN 115
AW++ + R L +FV + A V LL + V AV D+
Sbjct 113 AWAFLRISGRELWLTLLFVVEQVLAMIVVFIVALLIVILTAVVAVSGGDQG 163
> aly:ARALYDRAFT_495855 calmodulin-binding protein
Length=646
Score = 33.5 bits (75), Expect = 7.4, Method: Composition-based stats.
Identities = 17/44 (39%), Positives = 25/44 (57%), Gaps = 0/44 (0%)
Query 74 RPQQLNSIFVPQVDSANCSVAFDQLLCNVNFQVYAVQNLDRNGL 117
+PQ LNS Q + A+CS A DQ + N++ + N + NGL
Sbjct 454 QPQLLNSNPRAQFEVASCSTAQDQFMGNLHHTQSTIHNQNMNGL 497
> yel:LC20_03438 peptidase (caspase-1 like protein)
Length=328
Score = 33.5 bits (75), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 33/64 (52%), Gaps = 1/64 (2%)
Query 34 IDTVHAGFRAGAAYQSWVAPL-DGWNVLTSSGAWSYQSMKVRPQQLNSIFVPQVDSANCS 92
+D+ H+G G+A Q VA + DG +LT+S A Y + + S+FV + A +
Sbjct 126 LDSCHSGVAGGSALQQQVAEISDGVTILTASTAEQYATEENGAGVFTSLFVDALGGAAAN 185
Query 93 VAFD 96
+ D
Sbjct 186 LVGD 189
> dbr:Deba_0471 hypothetical protein
Length=830
Score = 33.5 bits (75), Expect = 7.7, Method: Composition-based stats.
Identities = 18/73 (25%), Positives = 34/73 (47%), Gaps = 0/73 (0%)
Query 31 KSNIDTVHAGFRAGAAYQSWVAPLDGWNVLTSSGAWSYQSMKVRPQQLNSIFVPQVDSAN 90
++ ++++ AG + GAA SW+ G L + W + ++ Q L + D+
Sbjct 410 RAGLESLSAGDQTGAAVLSWMTSDSGAGSLAAYAGWDTEDIRAMTQALGPMEAAMFDAQL 469
Query 91 CSVAFDQLLCNVN 103
S F Q L N++
Sbjct 470 ISDQFSQALDNIS 482
> mcc:719972 niban-like protein 2-like
Length=671
Score = 33.5 bits (75), Expect = 8.0, Method: Composition-based stats.
Identities = 17/45 (38%), Positives = 22/45 (49%), Gaps = 4/45 (9%)
Query 2 QSVPSLNLQNNPGRNVSGALGYNLRYWQWKSNIDTVHAGFRAGAA 46
+ +P+L Q PG SG R W W +D VHA AGA+
Sbjct 315 EQLPALRAQTLPGLRGSG----RARAWAWTELLDAVHAAVLAGAS 355
> ssc:100517808 FAM129C; family with sequence similarity 129,
member C
Length=618
Score = 33.5 bits (75), Expect = 8.9, Method: Composition-based stats.
Identities = 18/45 (40%), Positives = 21/45 (47%), Gaps = 4/45 (9%)
Query 2 QSVPSLNLQNNPGRNVSGALGYNLRYWQWKSNIDTVHAGFRAGAA 46
Q +P L Q PG SG R W W +D VHA AGA+
Sbjct 246 QLLPELRSQTLPGLRESG----RARAWSWTKLLDAVHAAVLAGAS 286
> mox:DAMO_3028 hypothetical protein
Length=438
Score = 33.1 bits (74), Expect = 9.7, Method: Composition-based stats.
Identities = 16/57 (28%), Positives = 28/57 (49%), Gaps = 1/57 (2%)
Query 28 WQWKSNIDTVHAGFRAGAAYQSWVAPLDGWNVLTSSGAWSYQSMKVRPQQLNSIFVP 84
W+W ++ GF + Y+ + AP G+ + GAW+ +S VR + F+P
Sbjct 369 WEWTASPFFSFPGFTVDSPYKEYSAPWFGYRKVLKGGAWATRSRLVR-NTFRNFFLP 424
Lambda K H a alpha
0.318 0.131 0.423 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 128610645009