bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-13_CDS_annotation_glimmer3.pl_2_9
Length=97
Score E
Sequences producing significant alignments: (Bits) Value
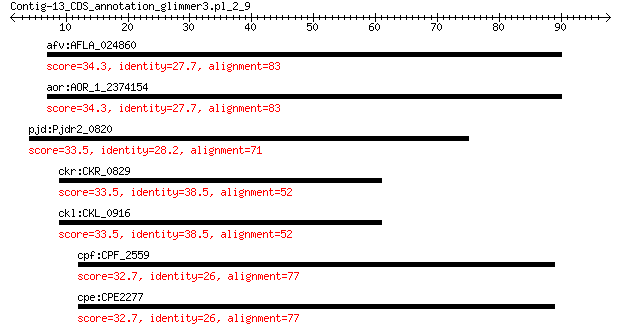
afv:AFLA_024860 MATH and UCH domain protein, putative 34.3 2.8
aor:AOR_1_2374154 AO090003001349; MATH and UCH domain protein 34.3 3.0
pjd:Pjdr2_0820 signal transduction histidine kinase LytS 33.5 4.3
ckr:CKR_0829 era; GTP-binding protein Era 33.5 4.8
ckl:CKL_0916 era; GTP-binding protein Era 33.5 4.8
cpf:CPF_2559 hypothetical protein 32.7 8.1
cpe:CPE2277 hypothetical protein 32.7 8.1
> afv:AFLA_024860 MATH and UCH domain protein, putative
Length=1270
Score = 34.3 bits (77), Expect = 2.8, Method: Composition-based stats.
Identities = 23/83 (28%), Positives = 38/83 (46%), Gaps = 4/83 (5%)
Query 7 RVKDLFQIIRTTTHEEQEEYVIVIGKHLATEARFSTREEAEQFIESKDYDLIWALIAALI 66
RV D Q++ T T E + + A T + D A+IAA+I
Sbjct 883 RVDDTAQVVTTETSSEHFDNNVTENSGEGAAASLETSHPENDIPNGAEND---AVIAAII 939
Query 67 DAKDVQEMEAQQEKDVQKVENVI 89
A+D+Q+ME +Q D+ ++ V+
Sbjct 940 -AEDIQQMETEQSTDIPQINEVM 961
> aor:AOR_1_2374154 AO090003001349; MATH and UCH domain protein
Length=1270
Score = 34.3 bits (77), Expect = 3.0, Method: Composition-based stats.
Identities = 23/83 (28%), Positives = 38/83 (46%), Gaps = 4/83 (5%)
Query 7 RVKDLFQIIRTTTHEEQEEYVIVIGKHLATEARFSTREEAEQFIESKDYDLIWALIAALI 66
RV D Q++ T T E + + A T + D A+IAA+I
Sbjct 883 RVDDTAQVVTTETSSEHFDNNVTENSGEGAAASLETSHPENDIPNGAEND---AVIAAII 939
Query 67 DAKDVQEMEAQQEKDVQKVENVI 89
A+D+Q+ME +Q D+ ++ V+
Sbjct 940 -AEDIQQMETEQSTDIPQINEVM 961
> pjd:Pjdr2_0820 signal transduction histidine kinase LytS
Length=575
Score = 33.5 bits (75), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 20/71 (28%), Positives = 40/71 (56%), Gaps = 2/71 (3%)
Query 4 REHRVKDLFQIIRTTTHEEQEEYVIVIGKHLATEARFSTREEAEQFIESKDYDLIWALIA 63
H + + F +IR+ T ++E V+ + HL+ R++TR E + ++ +L+ + +A
Sbjct 380 NPHFLYNCFSLIRSLTRLGEKESVMELALHLSRYYRYTTRSERQLAYLREEIELVKSYLA 439
Query 64 ALIDAKDVQEM 74
I A +VQ+M
Sbjct 440 --IQAMNVQDM 448
> ckr:CKR_0829 era; GTP-binding protein Era
Length=293
Score = 33.5 bits (75), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 20/54 (37%), Positives = 30/54 (56%), Gaps = 2/54 (4%)
Query 9 KDLFQIIRTTTHEEQEEYVIVIGKHLATEARFST--REEAEQFIESKDYDLIWA 60
K L+ I T E++ I+IGK A + ST RE+ E+F++SK Y +W
Sbjct 221 KGLYNIEATILCEKESHKGIIIGKKGAMLKKISTYAREDIEKFLDSKVYLEVWV 274
> ckl:CKL_0916 era; GTP-binding protein Era
Length=293
Score = 33.5 bits (75), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 20/54 (37%), Positives = 30/54 (56%), Gaps = 2/54 (4%)
Query 9 KDLFQIIRTTTHEEQEEYVIVIGKHLATEARFST--REEAEQFIESKDYDLIWA 60
K L+ I T E++ I+IGK A + ST RE+ E+F++SK Y +W
Sbjct 221 KGLYNIEATILCEKESHKGIIIGKKGAMLKKISTYAREDIEKFLDSKVYLEVWV 274
> cpf:CPF_2559 hypothetical protein
Length=873
Score = 32.7 bits (73), Expect = 8.1, Method: Composition-based stats.
Identities = 20/82 (24%), Positives = 39/82 (48%), Gaps = 5/82 (6%)
Query 12 FQIIRTTTHEEQEEYVIVIGKH--LATEARFSTR---EEAEQFIESKDYDLIWALIAALI 66
++I+ + + + V+ GK+ E RF T E E+ I+ K YDL + L I
Sbjct 523 YRIVSIDKNNDSNQEVLKEGKYGYFNVENRFITNISIENGEKLIQGKKYDLKYTLKPKSI 582
Query 67 DAKDVQEMEAQQEKDVQKVENV 88
KD+ + + + D ++ ++
Sbjct 583 ATKDISNISSDKLPDTIRLTDI 604
> cpe:CPE2277 hypothetical protein
Length=873
Score = 32.7 bits (73), Expect = 8.1, Method: Composition-based stats.
Identities = 20/82 (24%), Positives = 39/82 (48%), Gaps = 5/82 (6%)
Query 12 FQIIRTTTHEEQEEYVIVIGKH--LATEARFSTR---EEAEQFIESKDYDLIWALIAALI 66
++I+ + + + V+ GK+ E RF T E E+ I+ K YDL + L I
Sbjct 523 YRIVSIDKNNDSNQEVLKEGKYGYFNVENRFITNISIENGEKLIQGKKYDLKYTLKPKSI 582
Query 67 DAKDVQEMEAQQEKDVQKVENV 88
KD+ + + + D ++ ++
Sbjct 583 ATKDISNISSDKLPDTIRLTDI 604
Lambda K H a alpha
0.315 0.129 0.341 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 125035833240