bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-1_CDS_annotation_glimmer3.pl_2_1
Length=91
Score E
Sequences producing significant alignments: (Bits) Value
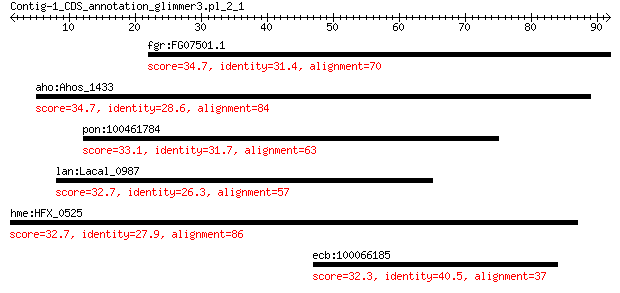
fgr:FG07501.1 hypothetical protein 34.7 1.3
aho:Ahos_1433 hypothetical protein 34.7 1.3
pon:100461784 uncharacterized LOC100461784 33.1 5.3
lan:Lacal_0987 proteptide peptidase M4 and M36 32.7 7.1
hme:HFX_0525 mutS; DNA mismatch repair protein 32.7 8.6
ecb:100066185 ABCA13; ATP-binding cassette, sub-family A (ABC1... 32.3 9.9
> fgr:FG07501.1 hypothetical protein
Length=265
Score = 34.7 bits (78), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 22/74 (30%), Positives = 38/74 (51%), Gaps = 4/74 (5%)
Query 22 ARAAAMSFVQER----ILTYRQAEELVRYLTNIHDPKNMWDGIWRIVSLPAGVSKSDFAA 77
RA A+SF +ER +L R A++L I D + +++++P +SK +
Sbjct 18 GRATAISFARERCLRLVLADRDADKLKDTQKTIIDMSKNQNQNIQVIAIPTDISKENDIE 77
Query 78 DLYDALCEEIGSVE 91
+LYD E+ G V+
Sbjct 78 NLYDTTIEKFGRVD 91
> aho:Ahos_1433 hypothetical protein
Length=195
Score = 34.7 bits (78), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 24/84 (29%), Positives = 38/84 (45%), Gaps = 9/84 (11%)
Query 5 ETEQLLRSQKFELTRQQARAAAMSFVQERILTYRQAEELVRYLTNIHDPKNMWDGIWRIV 64
+T L+SQ F L + ++E + E+LV NI PKN+ DGI +
Sbjct 74 KTLNYLKSQGFYLIPVHKQDLGDGLIKENTI-----EKLVNSYLNIVQPKNLADGIGLLN 128
Query 65 SLPAGVSKSDFAADLYDALCEEIG 88
+ AG + + D +CEE+
Sbjct 129 LISAGATITTQK----DGICEELN 148
> pon:100461784 uncharacterized LOC100461784
Length=419
Score = 33.1 bits (74), Expect = 5.3, Method: Composition-based stats.
Identities = 20/63 (32%), Positives = 34/63 (54%), Gaps = 4/63 (6%)
Query 12 SQKFELTRQQARAAAMSFVQERILTYRQAEELVRYLTNIHDPKNMWDGIWRIVSLPAGVS 71
++K + TR ++A ++ + + T+R + E +Y IHDP GIW L GVS
Sbjct 2 TKKLDDTRNTVKSANITSLNKEEKTFRNSRE--KYSLRIHDPNMKRIGIWPY--LETGVS 57
Query 72 KSD 74
K++
Sbjct 58 KNN 60
> lan:Lacal_0987 proteptide peptidase M4 and M36
Length=609
Score = 32.7 bits (73), Expect = 7.1, Method: Composition-based stats.
Identities = 15/58 (26%), Positives = 32/58 (55%), Gaps = 2/58 (3%)
Query 8 QLLRSQKFELTRQQARAAAMSFVQERILTY-RQAEELVRYLTNIHDPKNMWDGIWRIV 64
Q+ +++ FE+ + + A ++F +E + Y +Q ++ Y+ I +PK G W I+
Sbjct 107 QITQNEAFEIAKNEINAGEIAFKKEELFIYNKQETTILAYVVKI-EPKQTPIGSWEII 163
> hme:HFX_0525 mutS; DNA mismatch repair protein
Length=909
Score = 32.7 bits (73), Expect = 8.6, Method: Compositional matrix adjust.
Identities = 24/87 (28%), Positives = 46/87 (53%), Gaps = 2/87 (2%)
Query 1 MAQEETEQLLRSQKFELTRQQARAAAMSFVQERILTYRQAEELVRYLTNIHDPKNMWDG- 59
+A+E ++L ++L R +RAA+ S +L+ R +++ LT+ + + D
Sbjct 327 LARERVREVLDG-AYDLERLASRAASGSAGANDLLSVRDTLDVLPALTDAIEGTELADSP 385
Query 60 IWRIVSLPAGVSKSDFAADLYDALCEE 86
+ +VS P G + + A+L DAL E+
Sbjct 386 LAEVVSRPDGDAAAGLQAELADALAED 412
> ecb:100066185 ABCA13; ATP-binding cassette, sub-family A (ABC1),
member 13
Length=5059
Score = 32.3 bits (72), Expect = 9.9, Method: Composition-based stats.
Identities = 15/37 (41%), Positives = 22/37 (59%), Gaps = 1/37 (3%)
Query 47 LTNIHDPKNMWDGIWRIVSLPAGVSKSDFAADLYDAL 83
LT H P N+W + +V + +G+ KSD+ DL AL
Sbjct 2764 LTFAHHPDNLWKAVKTVVEVSSGM-KSDYGGDLSKAL 2799
Lambda K H a alpha
0.318 0.130 0.372 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 127599116940