bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-1_CDS_annotation_glimmer3.pl_2_3
Length=307
Score E
Sequences producing significant alignments: (Bits) Value
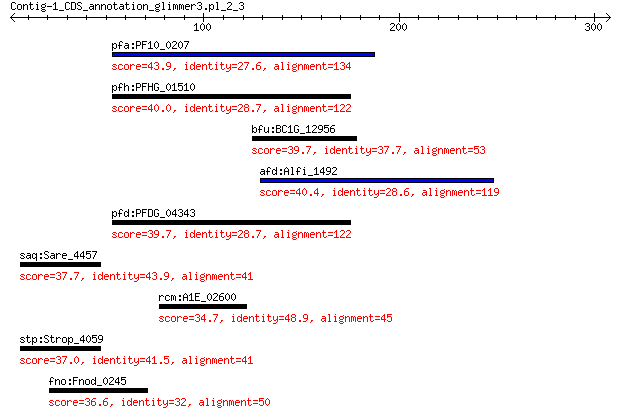
pfa:PF10_0207 conserved Plasmodium membrane protein 43.9 0.033
pfh:PFHG_01510 conserved hypothetical protein 40.0 0.40
bfu:BC1G_12956 hypothetical protein 39.7 0.41
afd:Alfi_1492 V8-like Glu-specific endopeptidase 40.4 0.48
pfd:PFDG_04343 conserved hypothetical protein 39.7 0.49
saq:Sare_4457 NADH dehydrogenase subunit G 37.7 3.7
rcm:A1E_02600 integrase catalytic subunit 34.7 5.5
stp:Strop_4059 NADH dehydrogenase subunit G (EC:1.6.5.3) 37.0 6.4
fno:Fnod_0245 small GTP-binding protein 36.6 6.5
> pfa:PF10_0207 conserved Plasmodium membrane protein
Length=454
Score = 43.9 bits (102), Expect = 0.033, Method: Compositional matrix adjust.
Identities = 37/139 (27%), Positives = 65/139 (47%), Gaps = 9/139 (6%)
Query 53 WFVRAHNIYKRLGYNLSNSYFCTFTL---KPEFYEAFCKEPYAFIRRFIDRMRKDQFLRY 109
+F ++ NI K + YN N F FT+ K ++ F K+ YAFI F ++ D+ ++
Sbjct 181 FFNKSSNISKLISYNSVNFLFLLFTILYFKSDYIYTFFKKKYAFIGEF-KNIKTDKLVKI 239
Query 110 RNPDTGRFCYRKISFPYLFVLEVADGKRAAQRRLHSEHRLHLHAIMFG--CPLPWWRVRH 167
T F + K F L +L + + ++L+ + L +H + F P++ V+
Sbjct 240 HTLSTLLFFFYK--FFVLRLLFILNSYNFFSQKLYLINNL-IHILFFSYISIFPYFLVQA 296
Query 168 YWMSFGLAWVSPLRHFGGV 186
W SF L + GG+
Sbjct 297 NWGSFHLLGYKKSKILGGL 315
> pfh:PFHG_01510 conserved hypothetical protein
Length=328
Score = 40.0 bits (92), Expect = 0.40, Method: Compositional matrix adjust.
Identities = 35/127 (28%), Positives = 61/127 (48%), Gaps = 9/127 (7%)
Query 53 WFVRAHNIYKRLGYNLSNSYFCTFTL---KPEFYEAFCKEPYAFIRRFIDRMRKDQFLRY 109
+F ++ NI K + YN N F FT+ K ++ F K+ YAFI F ++ D+ ++
Sbjct 198 FFNKSSNISKLISYNSVNFLFLLFTILYFKSDYIYTFFKKKYAFIGEF-KNIKTDKLVKI 256
Query 110 RNPDTGRFCYRKISFPYLFVLEVADGKRAAQRRLHSEHRLHLHAIMFG--CPLPWWRVRH 167
T F + K F L +L + + ++L+ + L +H + F P++ V+
Sbjct 257 HTLSTLLFFFYK--FFVLRLLFILNSYNFFSQKLYLINNL-IHILFFSYISIFPYFLVQA 313
Query 168 YWMSFGL 174
W SF L
Sbjct 314 NWGSFHL 320
> bfu:BC1G_12956 hypothetical protein
Length=234
Score = 39.7 bits (91), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 20/53 (38%), Positives = 28/53 (53%), Gaps = 3/53 (6%)
Query 125 PYLFVLEVADGKRAAQRRLHSEHRLHLHAIMFGCPLPWWRVRHYWMSFGLAWV 177
P++F L+ DGK A RR HA+ FGC L ++V+ W G+ WV
Sbjct 185 PWIF-LDQGDGK--ASRRGRKSTHARTHAVAFGCILVQFKVKKDWCDLGIIWV 234
> afd:Alfi_1492 V8-like Glu-specific endopeptidase
Length=704
Score = 40.4 bits (93), Expect = 0.48, Method: Compositional matrix adjust.
Identities = 34/129 (26%), Positives = 54/129 (42%), Gaps = 12/129 (9%)
Query 129 VLEVADGKRAAQRRLHSEHRLHLHAIMFGCP-LPWWRVRHYWMSFGLAWVSPLRHFGGVR 187
+LE+AD +R RR HRL++ +M P W ++ + V P G+R
Sbjct 519 LLELADAQRENNRRFKDGHRLYIAGLMRMQPDKAWASDANFTIRLTYGRVLPYDPADGIR 578
Query 188 YAMKYITKKSAVHWNDVPKE-----ILDLHGRLYVSHGFGRLSESEKDALRAYM----MT 238
Y Y T V + PK + D LY + FGR + +E + A++ +T
Sbjct 579 Y--NYYTTLKGVMEKEDPKNPTEFTVPDKLKELYAAKDFGRYANAEGELPVAFLADCDIT 636
Query 239 GCKQWFSVL 247
G V+
Sbjct 637 GGNSGSPVM 645
> pfd:PFDG_04343 conserved hypothetical protein
Length=311
Score = 39.7 bits (91), Expect = 0.49, Method: Compositional matrix adjust.
Identities = 35/127 (28%), Positives = 61/127 (48%), Gaps = 9/127 (7%)
Query 53 WFVRAHNIYKRLGYNLSNSYFCTFTL---KPEFYEAFCKEPYAFIRRFIDRMRKDQFLRY 109
+F ++ NI K + YN N F FT+ K ++ F K+ YAFI F ++ D+ ++
Sbjct 181 FFNKSSNISKLISYNSVNFLFLLFTILYFKSDYIYTFFKKKYAFIGEF-KNIKTDKLVKI 239
Query 110 RNPDTGRFCYRKISFPYLFVLEVADGKRAAQRRLHSEHRLHLHAIMFG--CPLPWWRVRH 167
T F + K F L +L + + ++L+ + L +H + F P++ V+
Sbjct 240 HTLSTLLFFFYK--FFVLRLLFILNSYNFFSQKLYLINNL-IHILFFSYISIFPYFLVQA 296
Query 168 YWMSFGL 174
W SF L
Sbjct 297 NWGSFHL 303
> saq:Sare_4457 NADH dehydrogenase subunit G
Length=839
Score = 37.7 bits (86), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 18/44 (41%), Positives = 27/44 (61%), Gaps = 6/44 (14%)
Query 6 LRITNPH---YIKLADQLGVEVPQFSNQSDYKLQVPCGKCVQCI 46
+ +T P I++A+QLGVE+P+F D+ L P G C QC+
Sbjct 18 IEVTAPKGALLIRVAEQLGVEIPRF---CDHPLLAPVGACRQCL 58
> rcm:A1E_02600 integrase catalytic subunit
Length=103
Score = 34.7 bits (78), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 22/49 (45%), Positives = 31/49 (63%), Gaps = 6/49 (12%)
Query 77 TLKPEFYE-AFCKEPYAFIRRFIDRMRKDQFL-RYRN--PDTGRFCYRK 121
TLK EFY+ AFCK+ Y + ++ DQ+L +Y N P +G+FCY K
Sbjct 28 TLKDEFYDIAFCKKIYNSLEDL--QIDLDQYLNKYNNTRPHSGKFCYGK 74
> stp:Strop_4059 NADH dehydrogenase subunit G (EC:1.6.5.3)
Length=839
Score = 37.0 bits (84), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 17/44 (39%), Positives = 27/44 (61%), Gaps = 6/44 (14%)
Query 6 LRITNPH---YIKLADQLGVEVPQFSNQSDYKLQVPCGKCVQCI 46
+ +T P I++A+Q+GVE+P+F D+ L P G C QC+
Sbjct 18 IEVTAPKGALLIRVAEQMGVEIPRF---CDHPLLAPVGACRQCL 58
> fno:Fnod_0245 small GTP-binding protein
Length=403
Score = 36.6 bits (83), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 16/50 (32%), Positives = 29/50 (58%), Gaps = 5/50 (10%)
Query 21 GVEVPQFSNQSDYKLQVPCGKCVQCIRKRQQHWFVRAHNIYKRLGYNLSN 70
GV++P++ +D KL + CG CV ++ +R ++KRLG ++N
Sbjct 334 GVDMPEYEEVADAKLVIHCGGCVNT-----RNQIMRRVRMFKRLGIPMTN 378
Lambda K H a alpha
0.328 0.141 0.480 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 527948001504