bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-22_CDS_annotation_glimmer3.pl_2_2
Length=561
Score E
Sequences producing significant alignments: (Bits) Value
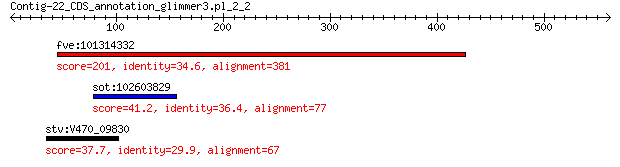
fve:101314332 capsid protein VP1-like 201 5e-55
sot:102603829 extensin-2-like 41.2 0.46
stv:V470_09830 capsid protein 37.7 6.5
> fve:101314332 capsid protein VP1-like
Length=421
Score = 201 bits (510), Expect = 5e-55, Method: Compositional matrix adjust.
Identities = 132/387 (34%), Positives = 201/387 (52%), Gaps = 48/387 (12%)
Query 45 VLPGDTFSVDTSAIIRMTTPKYPVMDDAFIDFYYFYCPNRILWDNFKQFMGEVEETPWMP 104
VLPGDTF+V+ + R+ TP +PVMD+ +D ++F+ PNR++W+N+ +FMGE ++ P
Sbjct 77 VLPGDTFNVNVTMFGRLATPIFPVMDNLHLDSFFFFVPNRLVWNNWVKFMGE-QDNP-AD 134
Query 105 KQEYSVPKIVINGKEKSPEPYEDSILDYMGIPTK----VKKVFKVNALPIRAYVKIWNEF 160
YS+P+ V + S+ DY G+PT V +ALP+RAY I+N++
Sbjct 135 SISYSIPQQVSPAGGYA----VGSLQDYFGLPTAGQVGVSNTVSHSALPVRAYNLIYNQW 190
Query 161 FRDENVDNAATIKTDDASIDYQDGNESEDNTDDILKKAIGGGRCLPVNKFHDYFTSCLPY 220
FRDEN+ N+ +D DG ++ +T+ L + K HDYFTS LP+
Sbjct 191 FRDENLQNSVV-------VDKGDGPDTTPSTNYTLLRR---------GKRHDYFTSALPW 234
Query 221 PQR-GPEVTLPMQGNAPVGMYKNDSLTEFGTINGNSEIFLNQALNGSALAPKISNSFKEG 279
PQ+ G V+LP+ +AP+ + S ++ G I+ + I N + G
Sbjct 235 PQKGGTAVSLPLGTSAPIA-FSGASGSDVGVIS-------------TTQGNLIKNMYSTG 280
Query 280 ARRALVTGSTNPTTQVSDAAYLAANLGETTATTVNDLRKAVAVQQYYEALARGGSRYREQ 339
+ +L GS T L A+L TA T+N LR++ +Q+ E ARGG+RY E
Sbjct 281 SGTSLKIGSATVATG------LYADLSAATAATINQLRQSFQIQKLLERDARGGTRYTEI 334
Query 340 VQALWDVVISDKTVQIPEYLGGGRYHVNMNQI-VQTSGQQSSTDTPIGETGAMSVTPVNE 398
+++ + V D +Q PEYLGGG +N+ I T TP G A
Sbjct 335 IRSHFGVASPDARLQRPEYLGGGSTPINIAPIAQTGGTGAQGTTTPQGNLAAFGTYMAKG 394
Query 399 SSFTKSFEEHGFIIGVCCVRHNHSYQQ 425
F++SF EHG +IG+ VR + +YQQ
Sbjct 395 HGFSQSFVEHGHVIGLVSVRADLTYQQ 421
> sot:102603829 extensin-2-like
Length=388
Score = 41.2 bits (95), Expect = 0.46, Method: Compositional matrix adjust.
Identities = 28/79 (35%), Positives = 43/79 (54%), Gaps = 3/79 (4%)
Query 79 FYCPNRILWDNFKQFMGEVEETPWMPKQEYSVPKIVINGKEKSPEPYEDSI--LDYMGIP 136
+Y + DN+K+ + V E P +PKQEY VP ++ N K P ED+ + Y+ +P
Sbjct 257 YYKKTSVPKDNYKK-VSSVSEAPLVPKQEYKVPYLLKNDYYKKPSIPEDNYRKVSYVPVP 315
Query 137 TKVKKVFKVNALPIRAYVK 155
+ K +KV +LP Y K
Sbjct 316 SVPKHEYKVPSLPKNDYYK 334
> stv:V470_09830 capsid protein
Length=427
Score = 37.7 bits (86), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 20/69 (29%), Positives = 39/69 (57%), Gaps = 2/69 (3%)
Query 35 GKLIPFYVDEVLPGDTFSVDTSAIIRMTTPKYPVMDDAFIDFYYFYCPNRILW-DNFKQF 93
G+LI V+ GD+F +D +R++ + + D+ +D + FY P+R ++ + + +F
Sbjct 26 GRLITISTTPVIAGDSFEMDAVGALRLSPLRRGLAIDSTVDIFTFYVPHRHVYGEQWIKF 85
Query 94 MGE-VEETP 101
M + V TP
Sbjct 86 MKDGVNATP 94
Lambda K H a alpha
0.316 0.133 0.397 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1271755612608