bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-27_CDS_annotation_glimmer3.pl_2_2
Length=107
Score E
Sequences producing significant alignments: (Bits) Value
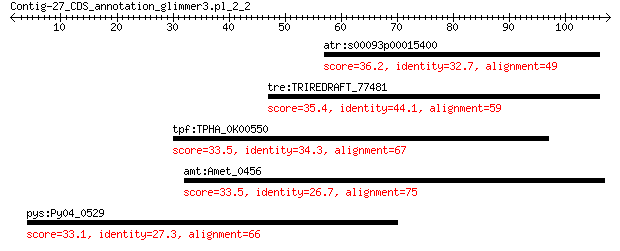
atr:s00093p00015400 AMTR_s00093p00015400; hypothetical protein 36.2 0.79
tre:TRIREDRAFT_77481 hypothetical protein 35.4 1.5
tpf:TPHA_0K00550 TPHA0K00550; hypothetical protein 33.5 5.1
amt:Amet_0456 sugar ABC transporter periplasmic protein-like p... 33.5 5.9
pys:Py04_0529 hypothetical protein 33.1 6.4
> atr:s00093p00015400 AMTR_s00093p00015400; hypothetical protein
Length=589
Score = 36.2 bits (82), Expect = 0.79, Method: Composition-based stats.
Identities = 16/51 (31%), Positives = 31/51 (61%), Gaps = 2/51 (4%)
Query 57 EFDIRT--DRFEIAIDAMDKINQSAASQIAKSSGETEAVKDFGTETKTDPE 105
EFD + + FE+A +++ + +I + +GETE ++++G +TDPE
Sbjct 7 EFDCESVIEAFEVATKDAERVQRETLRRILEENGETEYLQEWGLRGRTDPE 57
> tre:TRIREDRAFT_77481 hypothetical protein
Length=812
Score = 35.4 bits (80), Expect = 1.5, Method: Composition-based stats.
Identities = 26/66 (39%), Positives = 38/66 (58%), Gaps = 8/66 (12%)
Query 47 YTEKKDGVQP-EFDIR--TDRFEIAIDAMDKI----NQSAASQIAKSSGETEAVKDFGTE 99
Y EK + P E IR TDRF +AIDA+D++ N+ AA++ A + + EA ++ E
Sbjct 731 YKEKGNIDTPLELAIRNQTDRFSLAIDAIDRMPHLHNKGAAARQAMLNAQIEA-RNEAFE 789
Query 100 TKTDPE 105
DPE
Sbjct 790 KGMDPE 795
> tpf:TPHA_0K00550 TPHA0K00550; hypothetical protein
Length=424
Score = 33.5 bits (75), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 23/68 (34%), Positives = 35/68 (51%), Gaps = 5/68 (7%)
Query 30 IINGEANNMEDGVFPTIYTEKKDGVQPEFD-IRTDRFEIAIDAMDKINQSAASQIAKSSG 88
+++ E N GVF T Y + KD P+ D I + FE+ + K N S AS+IA S
Sbjct 240 VMSAEGNLYNPGVFNTDYIDDKDKTFPKVDKIAREYFEV----IKKCNGSHASRIAMKSH 295
Query 89 ETEAVKDF 96
+ ++ F
Sbjct 296 MFKILRPF 303
> amt:Amet_0456 sugar ABC transporter periplasmic protein-like
protein
Length=340
Score = 33.5 bits (75), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 20/75 (27%), Positives = 39/75 (52%), Gaps = 3/75 (4%)
Query 32 NGEANNMEDGVFPTIYTEKKDGVQPEFDIRTDRFEIAIDAMDKINQSAASQIAKSSGETE 91
+G N M+ G+ +Y DG +P ++ D F I + + +NQ A+++A +GE
Sbjct 107 SGAQNVMDAGIPLIVYDRLIDGFEPTMEVMGDNFTIGEETGEYLNQYFANELA--AGEVH 164
Query 92 AVKDFGTETKTDPEK 106
+ +F + T P++
Sbjct 165 -ILEFKGDNSTVPQQ 178
> pys:Py04_0529 hypothetical protein
Length=224
Score = 33.1 bits (74), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 18/71 (25%), Positives = 35/71 (49%), Gaps = 5/71 (7%)
Query 4 ATSRKGWINDPNLTYQAEPREVKLRKIIN-----GEANNMEDGVFPTIYTEKKDGVQPEF 58
+ + GWI + L+Y + P+ +R+ ++ + + E+ F YT+K G+ +
Sbjct 21 SMALAGWIGEYILSYSSLPKPKFIRRALSKLGFTYSSESYEENTFTMFYTKKNLGLSASW 80
Query 59 DIRTDRFEIAI 69
DI D+ I I
Sbjct 81 DIENDKLFINI 91
Lambda K H a alpha
0.308 0.127 0.348 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 125230604613