bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-28_CDS_annotation_glimmer3.pl_2_6
Length=301
Score E
Sequences producing significant alignments: (Bits) Value
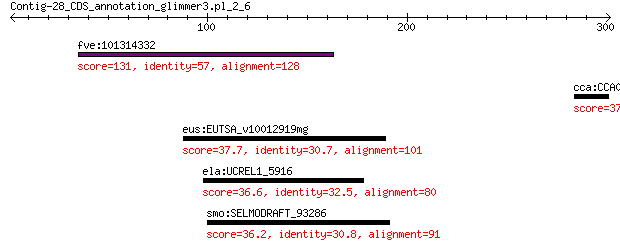
fve:101314332 capsid protein VP1-like 131 5e-32
cca:CCA00722 hypothetical protein 37.0 1.1
eus:EUTSA_v10012919mg hypothetical protein 37.7 2.9
ela:UCREL1_5916 putative carboxypeptidase s1 protein 36.6 7.1
smo:SELMODRAFT_93286 hypothetical protein 36.2 7.9
> fve:101314332 capsid protein VP1-like
Length=421
Score = 131 bits (329), Expect = 5e-32, Method: Compositional matrix adjust.
Identities = 73/131 (56%), Positives = 87/131 (66%), Gaps = 4/131 (3%)
Query 35 VIDNLWAVGDGVATATINQLRLAFQIQKLYEKDARGGTRYTEILRSHFGVTSPDSRLQRP 94
V L+A ATINQLR +FQIQKL E+DARGGTRYTEI+RSHFGV SPD+RLQRP
Sbjct 292 VATGLYADLSAATAATINQLRQSFQIQKLLERDARGGTRYTEIIRSHFGVASPDARLQRP 351
Query 95 EYLGGNRIPIRINQI--VQQSATQEGSTPQGNPVGL-SLTSDNHGDFTKSFTEHGFILGL 151
EYLGG PI I I + Q +TPQGN + + HG F++SF EHG ++GL
Sbjct 352 EYLGGGSTPINIAPIAQTGGTGAQGTTTPQGNLAAFGTYMAKGHG-FSQSFVEHGHVIGL 410
Query 152 MVARYDHTYQQ 162
+ R D TYQQ
Sbjct 411 VSVRADLTYQQ 421
> cca:CCA00722 hypothetical protein
Length=117
Score = 37.0 bits (84), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 13/17 (76%), Positives = 15/17 (88%), Gaps = 0/17 (0%)
Query 284 RCTRPMPMYSIPGLIDH 300
RC RPMP+YS+ GLIDH
Sbjct 100 RCARPMPVYSVSGLIDH 116
> eus:EUTSA_v10012919mg hypothetical protein
Length=652
Score = 37.7 bits (86), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 31/101 (31%), Positives = 45/101 (45%), Gaps = 9/101 (9%)
Query 88 DSRLQRPEYLGGNRIPIRINQIVQQSATQEGSTPQGNPVGLSLTSDNHGDFTKSFTEHGF 147
D+R P+ L GN + + +N I S QE T +G +T D+ DFT G
Sbjct 148 DTRKSHPDDLDGNHVGLNLNSI--NSVVQESLTGRG------ITIDSGVDFTAHVRYDGT 199
Query 148 ILGLMVARYDHTYQQGLDRMFSRKSRFDYYWPVFANIGEQA 188
L + V+R Y+Q + +FSR Y P +G A
Sbjct 200 FLSVYVSRNLEVYEQR-NLVFSRAIDLSAYLPETVYVGFTA 239
> ela:UCREL1_5916 putative carboxypeptidase s1 protein
Length=473
Score = 36.6 bits (83), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 26/80 (33%), Positives = 42/80 (53%), Gaps = 5/80 (6%)
Query 98 GGNRIPIRINQIVQQSATQEGSTPQGNPVGLSLTSDNHGDFTKSFTEHGFILGLMVARYD 157
GG P+ N+I++Q+ E T +G V L + N G + + TE + +GL+ Y
Sbjct 196 GGRYGPLFANRILEQNVAVEDGTVEGEHVNLVTLAMNDGLY--NMTE--WYIGLIEYSYS 251
Query 158 HTYQQGLDRMFSRKSRFDYY 177
+ Y+Q +D F R +DYY
Sbjct 252 NPYRQLIDDSF-RDELYDYY 270
> smo:SELMODRAFT_93286 hypothetical protein
Length=581
Score = 36.2 bits (82), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 28/96 (29%), Positives = 46/96 (48%), Gaps = 8/96 (8%)
Query 100 NRIPIRINQIVQQSATQEGSTPQGNPVGLSLTS-----DNHGDFTKSFTEHGFILGLMVA 154
+R I I +++++SA + S+ Q N G++ + NH D KS H F +
Sbjct 240 SREAIAIYELMEESARRSLSSLQPNADGVTFAAALGVCTNHVDRGKSL--HSFARDAGLD 297
Query 155 RYDHTYQQGLDRMFSRKSRFDYYWPVFANIGEQAVL 190
R ++ L M+SR R D W F I E++V+
Sbjct 298 R-ENAVGAALIAMYSRSGRLDEAWTAFEKIREKSVV 332
Lambda K H a alpha
0.317 0.134 0.404 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 509092715736