bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-2_CDS_annotation_glimmer3.pl_2_1
Length=206
Score E
Sequences producing significant alignments: (Bits) Value
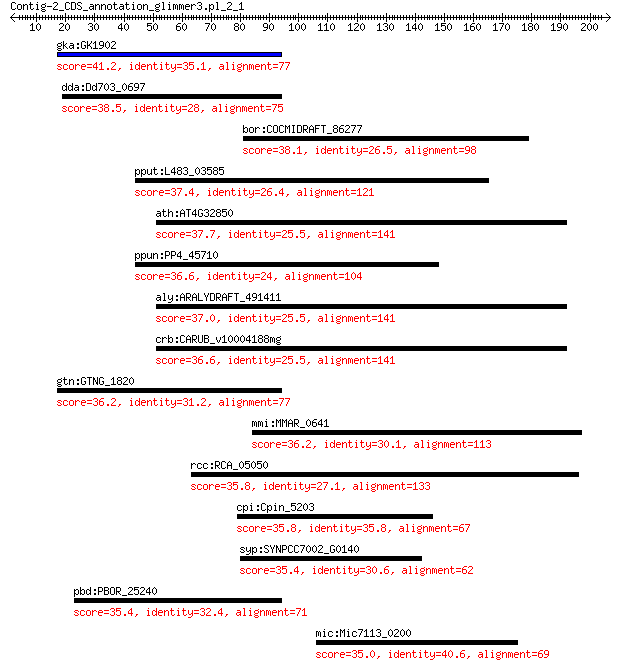
gka:GK1902 alpha-D-mannosidase (EC:3.2.1.24) 41.2 0.13
dda:Dd703_0697 glycoside hydrolase family protein 38.5 0.88
bor:COCMIDRAFT_86277 hypothetical protein 38.1 1.0
pput:L483_03585 iron dicitrate transporter FecR 37.4 1.6
ath:AT4G32850 nPAP; nuclear poly(a) polymerase 37.7 1.6
ppun:PP4_45710 putative transmembrane sensor 36.6 2.7
aly:ARALYDRAFT_491411 poly(A) polymerase 37.0 2.9
crb:CARUB_v10004188mg hypothetical protein 36.6 3.2
gtn:GTNG_1820 alpha-D-mannosidase 36.2 4.4
mmi:MMAR_0641 PPE family protein 36.2 4.6
rcc:RCA_05050 bifunctional N5-glutamine S-adenosyl-L-methionin... 35.8 4.9
cpi:Cpin_5203 TonB-dependent receptor 35.8 5.6
syp:SYNPCC7002_G0140 hypothetical protein 35.4 6.8
pbd:PBOR_25240 alpha-mannosidase 35.4 7.9
mic:Mic7113_0200 amino acid ABC transporter 35.0 9.1
> gka:GK1902 alpha-D-mannosidase (EC:3.2.1.24)
Length=1044
Score = 41.2 bits (95), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 27/79 (34%), Positives = 40/79 (51%), Gaps = 3/79 (4%)
Query 17 SGFDIGAKN--VFSAKCGELLPVYWDLGIPGCTYDIDIQYFTRTRPVQTAAYTRIREYFD 74
S +D+ AK + KCG +L V+ D + +DIDI Y + R V+ + I+E
Sbjct 724 SIYDLQAKREVLDDGKCGNVLQVFEDKPLRFDAWDIDIFYQEKKREVEDLQHVSIKEITS 783
Query 75 FYAVPIDLIWKSFDASVIQ 93
YAV I WK D+ + Q
Sbjct 784 LYAV-IHFEWKYMDSIIKQ 801
> dda:Dd703_0697 glycoside hydrolase family protein
Length=1037
Score = 38.5 bits (88), Expect = 0.88, Method: Compositional matrix adjust.
Identities = 21/75 (28%), Positives = 35/75 (47%), Gaps = 1/75 (1%)
Query 19 FDIGAKNVFSAKCGELLPVYWDLGIPGCTYDIDIQYFTRTRPVQTAAYTRIREYFDFYAV 78
FD + A CG +L VY D + +DI+I Y + RPV +RE +
Sbjct 720 FDRQHQRDVLADCGNVLTVYEDKPLKYDAWDIEIFYIQKQRPVTELLSATVREQGEL-CC 778
Query 79 PIDLIWKSFDASVIQ 93
I+ +W+ + ++Q
Sbjct 779 SIEFVWRYHHSRIVQ 793
> bor:COCMIDRAFT_86277 hypothetical protein
Length=333
Score = 38.1 bits (87), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 26/105 (25%), Positives = 46/105 (44%), Gaps = 7/105 (7%)
Query 81 DLIWKSFDASVIQMGETAP--VQAKDILTA-----LTVSGDLPYCSLSDLGLSCFFASGS 133
D++ ++ A+ + M P + + L + +T+ G L C L L A +
Sbjct 138 DVLEEAIPATAVHMTPMMPTKIHIRHSLESQKRKQITIPGSLSVCDDESLKLGTSLAQEN 197
Query 134 MSVPSLKSWQANNAYANIFGYIRGDVNYKLIHMLNYGNIIPNNMP 178
P L S + ++ A I G I+GD Y+ + YG I ++ P
Sbjct 198 SIKPDLTSGEEDDTQALIRGAIKGDAEYQTKLLRLYGEISASDGP 242
> pput:L483_03585 iron dicitrate transporter FecR
Length=316
Score = 37.4 bits (85), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 32/130 (25%), Positives = 51/130 (39%), Gaps = 11/130 (8%)
Query 44 PGCTYDIDIQYFTRTRPVQTAAYTRIREYFDFYA-VPIDLIWKSFDASVIQMGETAPVQA 102
P + Q++ + P A+ R+ + D A +P DL ++ D S Q ++
Sbjct 26 PTPRTEQQFQHWLASHPHNPLAWQRVSQLGDELAGLPKDLSRRTLDGS--QRQRSSRRDH 83
Query 103 KDILTALTVSGDLPYCSLSDLGLSCFFASGSMSVPSLKSWQA--------NNAYANIFGY 154
+L L VSG L + +GL A S + + WQA N A A Y
Sbjct 84 LKLLALLAVSGSLGWAMREPMGLPQLLADSSTATGERRQWQASDGSHIQLNTASAIDLHY 143
Query 155 IRGDVNYKLI 164
G +L+
Sbjct 144 SAGQRQLELV 153
> ath:AT4G32850 nPAP; nuclear poly(a) polymerase
Length=716
Score = 37.7 bits (86), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 36/153 (24%), Positives = 71/153 (46%), Gaps = 20/153 (13%)
Query 51 DIQYFTRTRPVQTAAYTRIREYFDFYAVPIDLIWKSFDASVI----QMGETAPVQAKDIL 106
+++ T PV A ++ F F +PIDL++ S V+ + ++ + D
Sbjct 141 EMEEVTELHPVPDAHVPVMK--FKFQGIPIDLLYASISLLVVPQDLDISSSSVLCEVDEP 198
Query 107 TALTVSGDLPYCSLSD--LGLSCFFASGSMSVPSLKSW-QANNAYANIFGYIRGDVNYKL 163
T +++G C ++D L L F ++ LK W + Y+N+ G++ G VN+ L
Sbjct 199 TVRSLNG----CRVADQILKLVPNFEHFRTTLRCLKYWAKKRGVYSNVTGFL-GGVNWAL 253
Query 164 IHMLNYGNIIPNNMPALNIGN-----SNYRWWN 191
+ + + PN +P++ + + +RW N
Sbjct 254 L-VARVCQLYPNAIPSMLVSRFFRVYTQWRWPN 285
> ppun:PP4_45710 putative transmembrane sensor
Length=316
Score = 36.6 bits (83), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 25/105 (24%), Positives = 45/105 (43%), Gaps = 3/105 (3%)
Query 44 PGCTYDIDIQYFTRTRPVQTAAYTRIREYFDFYA-VPIDLIWKSFDASVIQMGETAPVQA 102
P + Q++ + P A+ R+ + D A +P DL ++ D S Q ++
Sbjct 26 PSPRTEQQFQHWLASHPHNPLAWQRVSQLGDELAGLPKDLSRRTLDGS--QRQRSSRRDH 83
Query 103 KDILTALTVSGDLPYCSLSDLGLSCFFASGSMSVPSLKSWQANNA 147
+L+ L V G L + + LGL A S + + WQ ++
Sbjct 84 LKLLSLLAVGGSLGWATREPLGLPQLLADSSTATGERRQWQVSDG 128
> aly:ARALYDRAFT_491411 poly(A) polymerase
Length=748
Score = 37.0 bits (84), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 36/153 (24%), Positives = 71/153 (46%), Gaps = 20/153 (13%)
Query 51 DIQYFTRTRPVQTAAYTRIREYFDFYAVPIDLIWKSFDASVI----QMGETAPVQAKDIL 106
+++ T PV A ++ F F +PIDL++ S V+ + ++ + D
Sbjct 140 EMEEVTELHPVPDAHVPVMK--FKFQGIPIDLLYASISLLVVPQDLDISSSSVLCDVDEP 197
Query 107 TALTVSGDLPYCSLSD--LGLSCFFASGSMSVPSLKSW-QANNAYANIFGYIRGDVNYKL 163
T +++G C ++D L L F ++ LK W + Y+N+ G++ G VN+ L
Sbjct 198 TVRSLNG----CRVADQILKLVPNFEHFRTTLRCLKYWAKKRGVYSNVTGFL-GGVNWAL 252
Query 164 IHMLNYGNIIPNNMPALNIGN-----SNYRWWN 191
+ + + PN +P++ + + +RW N
Sbjct 253 L-VARVCQLYPNAIPSMLVSRFFRVYTQWRWPN 284
> crb:CARUB_v10004188mg hypothetical protein
Length=776
Score = 36.6 bits (83), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 36/153 (24%), Positives = 71/153 (46%), Gaps = 20/153 (13%)
Query 51 DIQYFTRTRPVQTAAYTRIREYFDFYAVPIDLIWKSFDASVI----QMGETAPVQAKDIL 106
+++ T PV A ++ F F +PIDL++ S V+ + ++ + D
Sbjct 141 EMEEVTELHPVPDAHVPVMK--FKFQGIPIDLLYASISLLVVPQDLDISSSSVLCDVDEP 198
Query 107 TALTVSGDLPYCSLSD--LGLSCFFASGSMSVPSLKSW-QANNAYANIFGYIRGDVNYKL 163
T +++G C ++D L L F ++ LK W + Y+N+ G++ G VN+ L
Sbjct 199 TVRSLNG----CRVADQILKLVPNFEHFRTTLRCLKYWAKKRGVYSNVTGFL-GGVNWAL 253
Query 164 IHMLNYGNIIPNNMPALNIGN-----SNYRWWN 191
+ + + PN +P++ + + +RW N
Sbjct 254 L-VARVCQLYPNAIPSMLVSRFFRVYTQWRWPN 285
> gtn:GTNG_1820 alpha-D-mannosidase
Length=1044
Score = 36.2 bits (82), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 24/79 (30%), Positives = 38/79 (48%), Gaps = 3/79 (4%)
Query 17 SGFDIGAKN--VFSAKCGELLPVYWDLGIPGCTYDIDIQYFTRTRPVQTAAYTRIREYFD 74
S +D AK + KCG +L V+ D + +DIDI Y + R V+ ++E
Sbjct 724 SIYDFEAKREVLDDGKCGNVLQVFEDKPLRFDAWDIDIFYQEKKREVEDLRRVSVKEITP 783
Query 75 FYAVPIDLIWKSFDASVIQ 93
YA+ + W D+S+ Q
Sbjct 784 LYAI-VHFEWGYMDSSIKQ 801
> mmi:MMAR_0641 PPE family protein
Length=2403
Score = 36.2 bits (82), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 34/123 (28%), Positives = 56/123 (46%), Gaps = 16/123 (13%)
Query 84 WKSFDASVIQMGETAP----VQAKDILTALTVSGDLPYCSLSDLG--LSCFFASGSMSVP 137
+ + ++ + +G + A D+LTA VSG +S+ G LS FF +G +
Sbjct 1044 YGTLESGIANLGNSVSGVFNTSALDLLTAANVSG------VSNFGSNLSGFFLAGDSADR 1097
Query 138 SLKSWQANNAYANIFGYIRGDVNYKLIHMLNY----GNIIPNNMPALNIGNSNYRWWNRE 193
AN+ NI GD N+ L ++ N GN+ N+ NIG++N+ + N
Sbjct 1098 IFNVGVANDGSFNIGSANLGDYNFGLGNVGNTNFGPGNLGSTNIGGANIGDTNFGFGNIG 1157
Query 194 AKK 196
A
Sbjct 1158 ANN 1160
> rcc:RCA_05050 bifunctional N5-glutamine S-adenosyl-L-methionine-dependent
methyltransferase/tRNA (m7G46) methyltransferase
Length=312
Score = 35.8 bits (81), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 36/140 (26%), Positives = 64/140 (46%), Gaps = 13/140 (9%)
Query 63 TAAYTRIREYFDFYAVPIDLIWKSFDASVIQMGETAPVQAKDILT---ALTVSGDLPYCS 119
TA T +RE F + +++ K+FD +++ ET+ ++A +L + V L +
Sbjct 178 TATPTLLREGF----IMSEVLEKTFDIEILKASETSTIKAPGMLAKHYSPKVPIRLNATN 233
Query 120 LSDLGLSCFFAS----GSMSVPSLKSWQANNAYANIFGYIRGDVNYKLIHMLNYGNIIPN 175
L D + F + G S+ A AN++ Y+R +Y H + Y + P
Sbjct 234 LEDKEIGLNFGNRNLMGRFSLNLSPKGDLIEAAANLYAYLRILDDYAASHDIEYIAVAP- 292
Query 176 NMPALNIGNSNYRWWNREAK 195
+P++NIG + R AK
Sbjct 293 -VPSVNIGAAINDRLKRAAK 311
> cpi:Cpin_5203 TonB-dependent receptor
Length=1004
Score = 35.8 bits (81), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 24/72 (33%), Positives = 36/72 (50%), Gaps = 6/72 (8%)
Query 79 PIDLIWKSFDASVIQMG-----ETAPVQAKDILTALTVSGDLPYCSLSDLGLSCFFASGS 133
P +WK+ DAS++ G T ++ KD +L V+G + DL +S + SGS
Sbjct 732 PTTTVWKNVDASIVNKGFEFTLGTTLIRTKDFSWSLDVNGATISNVIKDLPVSELY-SGS 790
Query 134 MSVPSLKSWQAN 145
+S P L AN
Sbjct 791 ISGPGLSGVNAN 802
> syp:SYNPCC7002_G0140 hypothetical protein
Length=533
Score = 35.4 bits (80), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 19/62 (31%), Positives = 34/62 (55%), Gaps = 4/62 (6%)
Query 80 IDLIWKSFDASVIQMGETAPVQAKDILTALTVSGDLPYCSLSDLGLSCFFASGSMSVPSL 139
ID++ K+ + S ET P K L A+ VSG + Y S+ + L+C + +M++ +
Sbjct 344 IDVLEKALEMSF----ETVPATGKTFLHAVDVSGSMSYYSVGSVHLTCAEIAATMALVTA 399
Query 140 KS 141
K+
Sbjct 400 KA 401
> pbd:PBOR_25240 alpha-mannosidase
Length=1061
Score = 35.4 bits (80), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 23/72 (32%), Positives = 36/72 (50%), Gaps = 2/72 (3%)
Query 23 AKNVFS-AKCGELLPVYWDLGIPGCTYDIDIQYFTRTRPVQTAAYTRIREYFDFYAVPID 81
A+ V + +CG LL V+ D +DID+ Y + R + R+ E AV ++
Sbjct 746 AREVLAPGECGNLLQVFEDKPKMYDAWDIDLFYQEKMREITDLQSVRLTESGPLQAV-LE 804
Query 82 LIWKSFDASVIQ 93
WK D+S+IQ
Sbjct 805 FKWKYMDSSIIQ 816
> mic:Mic7113_0200 amino acid ABC transporter
Length=520
Score = 35.0 bits (79), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 28/87 (32%), Positives = 39/87 (45%), Gaps = 18/87 (21%)
Query 106 LTALTVSGDLPYCSLSDLGLSCFFASGSMSVPSLKSWQANNAY------------ANIFG 153
+ AL ++ + Y L DLG + FFA G+ + L S Q N A+ A IFG
Sbjct 164 MLALGLNITVGYAGLLDLGYAAFFAIGAYTTGLLSSPQLNIAWNFWFVLPIAALVAAIFG 223
Query 154 YIRGDVNYKL------IHMLNYGNIIP 174
I G +L I L +G I+P
Sbjct 224 VILGSPTLRLRGDYLAIVTLGFGEIVP 250
Lambda K H a alpha
0.321 0.137 0.436 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 235643300514