bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
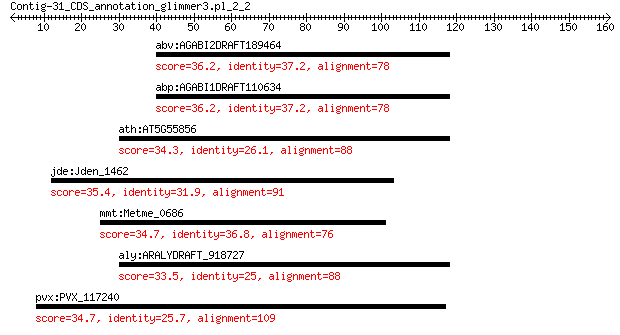
Query= Contig-31_CDS_annotation_glimmer3.pl_2_2
Length=160
Score E
Sequences producing significant alignments: (Bits) Value
abv:AGABI2DRAFT189464 AGABI2DRAFT_189464; hypothetical protein 36.2 2.7
abp:AGABI1DRAFT110634 AGABI1DRAFT_110634; hypothetical protein 36.2 2.7
ath:AT5G55856 putative small ubiquitin-related modifier 8 34.3
jde:Jden_1462 aldo/keto reductase 35.4 3.8
mmt:Metme_0686 hypothetical protein 34.7 5.7
aly:ARALYDRAFT_918727 hypothetical protein 33.5 8.2
pvx:PVX_117240 hypothetical protein 34.7 8.3
> abv:AGABI2DRAFT189464 AGABI2DRAFT_189464; hypothetical protein
Length=985
Score = 36.2 bits (82), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 29/86 (34%), Positives = 44/86 (51%), Gaps = 9/86 (10%)
Query 40 YIRVRSDISMLFHAEATAKKIGTEGFRFLAESRRVKSSPTQAMMDKMPD-----DLILDT 94
Y RVR I F A AKK +EG++ LAE V P+ A+ ++ P+ +LIL T
Sbjct 858 YNRVRQAICSGFFRHA-AKKDPSEGYKTLAEGTPVYIHPSSALFNRNPEWLVYHELILTT 916
Query 95 LKSRH---LQQPSELLAFSEQLSALA 117
+ H + +P L+ F+ Q +A
Sbjct 917 REYCHNVTVIEPKWLVEFAPQCFKVA 942
> abp:AGABI1DRAFT110634 AGABI1DRAFT_110634; hypothetical protein
Length=985
Score = 36.2 bits (82), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 29/86 (34%), Positives = 44/86 (51%), Gaps = 9/86 (10%)
Query 40 YIRVRSDISMLFHAEATAKKIGTEGFRFLAESRRVKSSPTQAMMDKMPD-----DLILDT 94
Y RVR I F A AKK +EG++ LAE V P+ A+ ++ P+ +LIL T
Sbjct 858 YNRVRQAICSGFFRHA-AKKDPSEGYKTLAEGTPVYIHPSSALFNRNPEWLVYHELILTT 916
Query 95 LKSRH---LQQPSELLAFSEQLSALA 117
+ H + +P L+ F+ Q +A
Sbjct 917 REYCHNVTVIEPKWLVEFAPQCFKVA 942
> ath:AT5G55856 putative small ubiquitin-related modifier 8
Length=97
Score = 34.3 bits (77), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 23/90 (26%), Positives = 43/90 (48%), Gaps = 17/90 (19%)
Query 30 QVEHTDPLESYIRVRSDISMLFHAEATAKKIGTE--GFRFLAESRRVKSSPTQAMMDKMP 87
+V++ D + Y R++ D+ + A + K+G E RFL + R+K +++ P
Sbjct 18 KVKNQDDICVYFRIKRDVELRKMMHAYSDKVGVEMSTLRFLFDGNRIK-------LNQTP 70
Query 88 DDLILDTLKSRHLQQPSELLAFSEQLSALA 117
++L L+ E+ AF EQL +
Sbjct 71 NEL--------GLEDEDEIEAFGEQLGGFS 92
> jde:Jden_1462 aldo/keto reductase
Length=333
Score = 35.4 bits (80), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 29/100 (29%), Positives = 52/100 (52%), Gaps = 10/100 (10%)
Query 12 VEGEIT-LEVDNVDLFRIEQVEHTDPLESYIRVRSDIS------MLFHAEATAKKIGTEG 64
+ G +T L D VDL++ + +H+ PLE + +DI + +E TA++I TEG
Sbjct 104 INGSLTRLGTDYVDLYQAHRYDHSTPLEETMIAFADIVRSGKALYIGVSEWTAEQI-TEG 162
Query 65 FRFLAES--RRVKSSPTQAMMDKMPDDLILDTLKSRHLQQ 102
R+ E + + S P M+ ++ +D ++ T + L Q
Sbjct 163 HRYAKELGFQLISSQPQYNMLWRVIEDKVVPTSQELGLSQ 202
> mmt:Metme_0686 hypothetical protein
Length=187
Score = 34.7 bits (78), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 28/78 (36%), Positives = 38/78 (49%), Gaps = 9/78 (12%)
Query 25 LFRIEQVEHTDPLESYIRVRSDISMLFHAEATAKKIGTEGF--RFLAESRRVKSSPTQAM 82
LF ++QVE + LES I+ SDI H T I +GF R A P QA+
Sbjct 4 LFTLQQVE-LEQLESLIKDPSDIFFFLHGPET--YIPKKGFFSRLFAAPEPPAQRPWQAL 60
Query 83 MDKMPDDLILDTLKSRHL 100
P+D +LD K+ H+
Sbjct 61 ----PEDSLLDLDKNWHI 74
> aly:ARALYDRAFT_918727 hypothetical protein
Length=131
Score = 33.5 bits (75), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 22/90 (24%), Positives = 42/90 (47%), Gaps = 17/90 (19%)
Query 30 QVEHTDPLESYIRVRSDISMLFHAEATAKKIGTE--GFRFLAESRRVKSSPTQAMMDKMP 87
+++ D + Y R++ D+ + A + K+G E RFL + R+K +++ P
Sbjct 52 HIKNQDDICVYFRIKRDVELRKMMHAYSAKVGVEMSTLRFLFDGNRIK-------LNQTP 104
Query 88 DDLILDTLKSRHLQQPSELLAFSEQLSALA 117
++L L+ E+ AF EQL +
Sbjct 105 NEL--------GLEDEDEIEAFGEQLGGFS 126
> pvx:PVX_117240 hypothetical protein
Length=2952
Score = 34.7 bits (78), Expect = 8.3, Method: Composition-based stats.
Identities = 28/110 (25%), Positives = 49/110 (45%), Gaps = 6/110 (5%)
Query 8 NPAVVEGEITLEVDNVDLFRIEQVEHTDPLESY-IRVRSDISMLFHAEATAKKIGTEGFR 66
N A G + DN +++++ ++P E+Y I +RS I+ML + T + +G+E
Sbjct 2440 NSADENGHGSTSSDNNKTGTVKKLQESEPNENYFIAIRSKINMLGSSNRTNEAVGSE--- 2496
Query 67 FLAESRRVKSSPTQAMMDKMPDDLILDTLKSRHLQQPSELLAFSEQLSAL 116
+ P Q K PD IL +++++ S S S L
Sbjct 2497 --ENPPEQRDLPAQVKKKKKPDATILSDPENKYVTNKSSASKHSRSRSKL 2544
Lambda K H a alpha
0.312 0.126 0.336 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 128616150254