bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-34_CDS_annotation_glimmer3.pl_2_1
Length=184
Score E
Sequences producing significant alignments: (Bits) Value
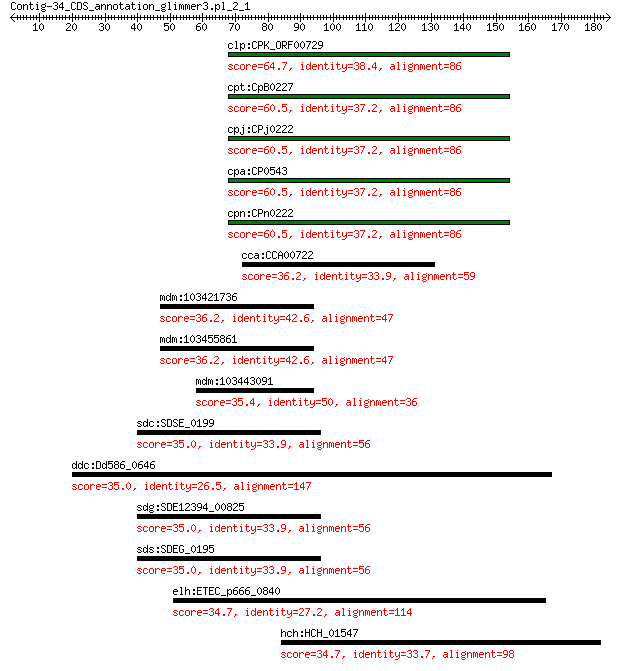
clp:CPK_ORF00729 hypothetical protein 64.7 7e-11
cpt:CpB0227 hypothetical protein 60.5 2e-09
cpj:CPj0222 hypothetical protein 60.5 2e-09
cpa:CP0543 hypothetical protein 60.5 2e-09
cpn:CPn0222 hypothetical protein 60.5 2e-09
cca:CCA00722 hypothetical protein 36.2 1.2
mdm:103421736 probable xyloglucan glycosyltransferase 9 36.2
mdm:103455861 probable xyloglucan glycosyltransferase 12 36.2
mdm:103443091 probable xyloglucan glycosyltransferase 12 35.4
sdc:SDSE_0199 Anaerobic sulfatase-maturating enzyme ydeM 35.0 7.0
ddc:Dd586_0646 oxidoreductase molybdopterin-binding protein 35.0 7.1
sdg:SDE12394_00825 putative transcriptional regulator 35.0 7.4
sds:SDEG_0195 transcriptional regulator 35.0 7.5
elh:ETEC_p666_0840 putative Oxidoreductase, molybdopterin binding 34.7 9.0
hch:HCH_01547 hypothetical protein 34.7 9.7
> clp:CPK_ORF00729 hypothetical protein
Length=121
Score = 64.7 bits (156), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 33/87 (38%), Positives = 52/87 (60%), Gaps = 6/87 (7%)
Query 68 FLIVPCHKCPLCNEKKAQQWSFRALCESYTSNKQAYFITLTYNNEHLPKNGVFPE-EIQL 126
++++PC KC C + A+ WS+R + E+ K F+TLTY+++HLP+ G + +QL
Sbjct 19 WVLMPCLKCRFCRTQHAKVWSYRCVHEASLYEKNC-FLTLTYDDKHLPQYGSLVKLHLQL 77
Query 127 FFKRLRTKLDRRGISHNLRYIAVSEYG 153
F KRLR ++ H +RY EYG
Sbjct 78 FLKRLRDRIS----PHKIRYFGCGEYG 100
> cpt:CpB0227 hypothetical protein
Length=113
Score = 60.5 bits (145), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 32/87 (37%), Positives = 50/87 (57%), Gaps = 6/87 (7%)
Query 68 FLIVPCHKCPLCNEKKAQQWSFRALCESYTSNKQAYFITLTYNNEHLPKNGVFPE-EIQL 126
++++PC KC C + A+ WS+R + E+ K F+TLTY+++HLP+ G + +QL
Sbjct 19 WVLMPCLKCRFCRTQHAKVWSYRCVHEASLYEKNC-FLTLTYDDKHLPQYGSLVKLHLQL 77
Query 127 FFKRLRTKLDRRGISHNLRYIAVSEYG 153
F KRLR + H +RY YG
Sbjct 78 FLKRLRKMIS----PHKIRYFECGAYG 100
> cpj:CPj0222 hypothetical protein
Length=113
Score = 60.5 bits (145), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 32/87 (37%), Positives = 50/87 (57%), Gaps = 6/87 (7%)
Query 68 FLIVPCHKCPLCNEKKAQQWSFRALCESYTSNKQAYFITLTYNNEHLPKNGVFPE-EIQL 126
++++PC KC C + A+ WS+R + E+ K F+TLTY+++HLP+ G + +QL
Sbjct 19 WVLMPCLKCRFCRTQHAKVWSYRCVHEASLYEKNC-FLTLTYDDKHLPQYGSLVKLHLQL 77
Query 127 FFKRLRTKLDRRGISHNLRYIAVSEYG 153
F KRLR + H +RY YG
Sbjct 78 FLKRLRKMIS----PHKIRYFECGAYG 100
> cpa:CP0543 hypothetical protein
Length=113
Score = 60.5 bits (145), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 32/87 (37%), Positives = 50/87 (57%), Gaps = 6/87 (7%)
Query 68 FLIVPCHKCPLCNEKKAQQWSFRALCESYTSNKQAYFITLTYNNEHLPKNGVFPE-EIQL 126
++++PC KC C + A+ WS+R + E+ K F+TLTY+++HLP+ G + +QL
Sbjct 19 WVLMPCLKCRFCRTQHAKVWSYRCVHEASLYEKNC-FLTLTYDDKHLPQYGSLVKLHLQL 77
Query 127 FFKRLRTKLDRRGISHNLRYIAVSEYG 153
F KRLR + H +RY YG
Sbjct 78 FLKRLRKMIS----PHKIRYFECGAYG 100
> cpn:CPn0222 hypothetical protein
Length=113
Score = 60.5 bits (145), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 32/87 (37%), Positives = 50/87 (57%), Gaps = 6/87 (7%)
Query 68 FLIVPCHKCPLCNEKKAQQWSFRALCESYTSNKQAYFITLTYNNEHLPKNGVFPE-EIQL 126
++++PC KC C + A+ WS+R + E+ K F+TLTY+++HLP+ G + +QL
Sbjct 19 WVLMPCLKCRFCRTQHAKVWSYRCVHEASLYEKNC-FLTLTYDDKHLPQYGSLVKLHLQL 77
Query 127 FFKRLRTKLDRRGISHNLRYIAVSEYG 153
F KRLR + H +RY YG
Sbjct 78 FLKRLRKMIS----PHKIRYFECGAYG 100
> cca:CCA00722 hypothetical protein
Length=117
Score = 36.2 bits (82), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 20/60 (33%), Positives = 34/60 (57%), Gaps = 2/60 (3%)
Query 72 PCHKCPLCNEKKAQQWSFRALCESYTSNKQAYFITLTYNNEHLPKNG-VFPEEIQLFFKR 130
P HK + A+ WS+R + E+ + Q F+TLTY + +LP+ G + +++LF R
Sbjct 31 PMHKNEVLVHGSAKVWSYRCIHEA-SLYGQNSFLTLTYEDRNLPEKGSLVRRDVRLFLMR 89
> mdm:103421736 probable xyloglucan glycosyltransferase 9
Length=818
Score = 36.2 bits (82), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 20/49 (41%), Positives = 28/49 (57%), Gaps = 6/49 (12%)
Query 47 NKNNIDKYTIVNPVTGET--FPMFLIVPCHKCPLCNEKKAQQWSFRALC 93
N + K ++P TGET FPM L+ + P+CNEK+ Q S A+C
Sbjct 339 NIKPVPKEGALDPETGETGYFPMVLV----QIPMCNEKEVYQTSIAAVC 383
> mdm:103455861 probable xyloglucan glycosyltransferase 12
Length=700
Score = 36.2 bits (82), Expect = 4.1, Method: Composition-based stats.
Identities = 20/49 (41%), Positives = 27/49 (55%), Gaps = 6/49 (12%)
Query 47 NKNNIDKYTIVNPVTGET--FPMFLIVPCHKCPLCNEKKAQQWSFRALC 93
N + K +P TGET FPM L+ + P+CNEK+ Q S A+C
Sbjct 221 NIKPVPKEXAXDPETGETGYFPMVLV----QIPMCNEKEVYQTSIAAVC 265
> mdm:103443091 probable xyloglucan glycosyltransferase 12
Length=699
Score = 35.4 bits (80), Expect = 6.3, Method: Composition-based stats.
Identities = 18/38 (47%), Positives = 24/38 (63%), Gaps = 6/38 (16%)
Query 58 NPVTGET--FPMFLIVPCHKCPLCNEKKAQQWSFRALC 93
+P TGET FPM L+ + P+CNEK+ Q S A+C
Sbjct 231 DPETGETGYFPMVLV----QIPMCNEKEVYQTSIAAVC 264
> sdc:SDSE_0199 Anaerobic sulfatase-maturating enzyme ydeM
Length=372
Score = 35.0 bits (79), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 19/56 (34%), Positives = 28/56 (50%), Gaps = 2/56 (4%)
Query 40 HVNKNHVNKNNIDKYTIVNPVTGETFPMFLIVPCHKCPLCNEKKAQQWSFRALCES 95
++N VN+N IDK+ I + P LI CP+ K+ SF+ +CES
Sbjct 294 NINTGFVNQNKIDKWVINVDDSCSDCPYLLICKSGNCPMV--KRINGVSFKRICES 347
> ddc:Dd586_0646 oxidoreductase molybdopterin-binding protein
Length=256
Score = 35.0 bits (79), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 39/155 (25%), Positives = 68/155 (44%), Gaps = 23/155 (15%)
Query 20 LVQKYRTFTMPSGTYHGSVLHVNKNHVNKNNIDKYTIVNPVTGETFPMFLIVPCHKCPLC 79
LV+ R +T+P V + + H+ ++ ++ V G F FL + +
Sbjct 117 LVRDTRRWTLPELYRLQQVSQITR-HIC---VEGWSAVGKWGGVPFHHFLAL------IG 166
Query 80 NEKKAQQWSFRALCESYTSNKQAY------FITLTYNNEHLPKNGVFPEEIQLFFKRLRT 133
+K+AQ +FR + YTS A + LT++N+ LP FP +I R+ T
Sbjct 167 ADKRAQYVNFRCADDYYTSIDMATALHPQTLLALTWDNQILPPEYGFPMKI-----RIPT 221
Query 134 KLDRRGISHNLRYIAVSEY--GHWSKRRVHSFGRF 166
KL + H L + + G+W + + FG +
Sbjct 222 KLGYKNPKHVLSIEVTNRFLGGYWEDQGYNWFGGY 256
> sdg:SDE12394_00825 putative transcriptional regulator
Length=429
Score = 35.0 bits (79), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 19/56 (34%), Positives = 28/56 (50%), Gaps = 2/56 (4%)
Query 40 HVNKNHVNKNNIDKYTIVNPVTGETFPMFLIVPCHKCPLCNEKKAQQWSFRALCES 95
++N VN+N IDK+ I + P LI CP+ K+ SF+ +CES
Sbjct 351 NINTGFVNQNKIDKWVINVDDSCSDCPYLLICKSGNCPMV--KRINGVSFKRICES 404
> sds:SDEG_0195 transcriptional regulator
Length=437
Score = 35.0 bits (79), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 19/56 (34%), Positives = 28/56 (50%), Gaps = 2/56 (4%)
Query 40 HVNKNHVNKNNIDKYTIVNPVTGETFPMFLIVPCHKCPLCNEKKAQQWSFRALCES 95
++N VN+N IDK+ I + P LI CP+ K+ SF+ +CES
Sbjct 359 NINTGFVNQNKIDKWVINVDDSCSDCPYLLICKSGNCPMV--KRINGVSFKRICES 412
> elh:ETEC_p666_0840 putative Oxidoreductase, molybdopterin binding
Length=256
Score = 34.7 bits (78), Expect = 9.0, Method: Compositional matrix adjust.
Identities = 31/123 (25%), Positives = 59/123 (48%), Gaps = 21/123 (17%)
Query 51 IDKYTIVNPVTGETFPMFLIVPCHKCPLCNEKKAQQWSFRALCESYTSNKQAY------F 104
++ ++ + G F FL + + + +A+ SFR + + YTS A
Sbjct 144 VEGWSAIGEWGGVPFRHFLTL------IGADMRAKYVSFRCMDDYYTSIDMATALHPQTL 197
Query 105 ITLTYNNEHLPKNGVFPEEIQLFFKRLRTKLDRRGISHNLRYIAVSEY---GHWSKRRVH 161
+TLT+NN+ LPK +P ++ R+ TKL + H ++ I ++ + G+W + +
Sbjct 198 LTLTWNNQILPKEYGYPMKL-----RIPTKLGYKNPKH-VQVIEITNHFPGGYWEDQGYN 251
Query 162 SFG 164
FG
Sbjct 252 WFG 254
> hch:HCH_01547 hypothetical protein
Length=373
Score = 34.7 bits (78), Expect = 9.7, Method: Compositional matrix adjust.
Identities = 33/113 (29%), Positives = 55/113 (49%), Gaps = 17/113 (15%)
Query 84 AQQWSFRALCESYTSNKQAYFITLTYNNEHLPKNGVFPEEIQLFFKRLRTKLD------- 136
AQ S + + ESY S K+ + LT ++ +P+NG+ ++I+ +++ T+L+
Sbjct 138 AQNKSAQFVGESYASQKKYFDFKLT--SKGMPRNGLVEDKIRAAAQQVYTQLEAYLAPER 195
Query 137 RRGISHNLR-YIAVSEYGHWSKRRVHSFGRFFTFSH-------IFHDIIDTTV 181
RR I NL Y +EY K+ F FSH + D ++TTV
Sbjct 196 RRQIILNLELYGDRAEYEALRKKVFPGFNAAAFFSHGANTIYMVNEDQLETTV 248
Lambda K H a alpha
0.324 0.137 0.445 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 172592981444