bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-34_CDS_annotation_glimmer3.pl_2_4
Length=374
Score E
Sequences producing significant alignments: (Bits) Value
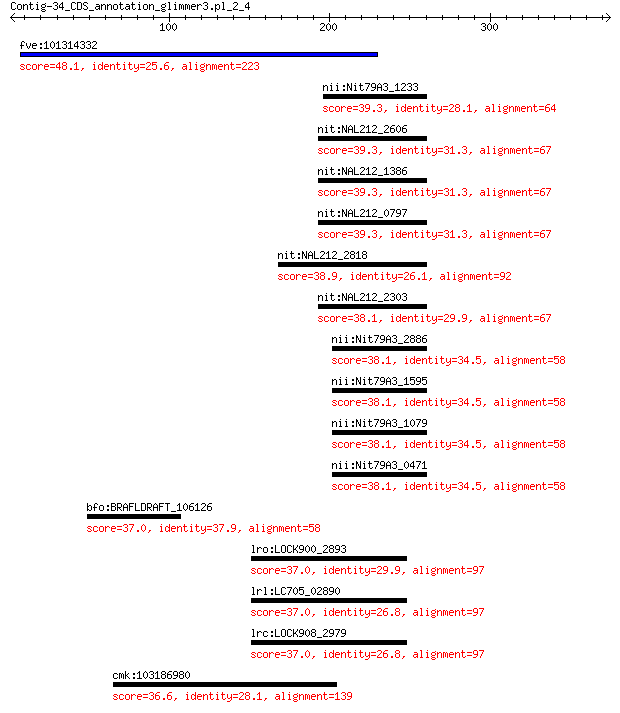
fve:101314332 capsid protein VP1-like 48.1 0.002
nii:Nit79A3_1233 methane monooxygenase/ammonia monooxygenase s... 39.3 1.0
nit:NAL212_2606 methane monooxygenase/ammonia monooxygenase su... 39.3 1.0
nit:NAL212_1386 methane monooxygenase/ammonia monooxygenase su... 39.3 1.0
nit:NAL212_0797 methane monooxygenase/ammonia monooxygenase su... 39.3 1.0
nit:NAL212_2818 methane monooxygenase/ammonia monooxygenase su... 38.9 1.3
nit:NAL212_2303 methane monooxygenase/ammonia monooxygenase su... 38.1 2.1
nii:Nit79A3_2886 methane monooxygenase/ammonia monooxygenase s... 38.1 2.2
nii:Nit79A3_1595 methane monooxygenase/ammonia monooxygenase s... 38.1 2.2
nii:Nit79A3_1079 methane monooxygenase/ammonia monooxygenase s... 38.1 2.2
nii:Nit79A3_0471 methane monooxygenase/ammonia monooxygenase s... 38.1 2.2
bfo:BRAFLDRAFT_106126 hypothetical protein 37.0 6.8
lro:LOCK900_2893 Hypothetical protein 37.0 7.8
lrl:LC705_02890 adhesion exoprotein 37.0 8.1
lrc:LOCK908_2979 Adhesion exoprotein 37.0 8.2
cmk:103186980 cops2; COP9 signalosome subunit 2 36.6
> fve:101314332 capsid protein VP1-like
Length=421
Score = 48.1 bits (113), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 57/233 (24%), Positives = 94/233 (40%), Gaps = 42/233 (18%)
Query 7 NTTYSLHQCNWERDFLTTAVPNPQQGANAPLVGLTVGDVVTRA-------DDGTYSIQKQ 59
+T Y+L + D+ T+A+P PQ+G A V L +G A D G S +
Sbjct 213 STNYTLLRRGKRHDYFTSALPWPQKGGTA--VSLPLGTSAPIAFSGASGSDVGVISTTQG 270
Query 60 TVLVDEDGSKYGISYKVSEDGESLVGVDYDPVSEKTPVTAINSYAELAALTTTEGSGFTI 119
++ + + G S K+ + A YA+L+A T TI
Sbjct 271 NLIKNMYSTGSGTSLKIG-----------------SATVATGLYADLSAATAA-----TI 308
Query 120 ETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSMRTVEQ 179
LR QK LE + R G Y +I++ + + L PE++GG S +++ + Q
Sbjct 309 NQLRQSFQIQKLLERDARGGTRYTEIIRSHFGVASPDARLQRPEYLGGGSTPINIAPIAQ 368
Query 180 TVDQQSTSS---QGQYAEALGSKSGIAGVYGSTSNNIEVFCDEESYIIGLLTV 229
T + + QG A G Y + + E ++IGL++V
Sbjct 369 TGGTGAQGTTTPQGNLAA--------FGTYMAKGHGFSQSFVEHGHVIGLVSV 413
> nii:Nit79A3_1233 methane monooxygenase/ammonia monooxygenase
subunit C
Length=269
Score = 39.3 bits (90), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 18/64 (28%), Positives = 35/64 (55%), Gaps = 0/64 (0%)
Query 196 LGSKSGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPE 255
+ + G G S + ++ ++ D + Y IG+LT+ V I+ + F Y+ +D +PE
Sbjct 1 MATTYGTTGASSSANYDMSLWYDSKYYKIGMLTMLLVAIFWIWYQRTFAYSHGMDSMEPE 60
Query 256 FDRI 259
FD++
Sbjct 61 FDKV 64
> nit:NAL212_2606 methane monooxygenase/ammonia monooxygenase
subunit C
Length=269
Score = 39.3 bits (90), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 21/67 (31%), Positives = 37/67 (55%), Gaps = 4/67 (6%)
Query 193 AEALGSKSGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHY 252
A G+ SG A S + ++ ++ D + Y IG+LT+ V I+ + F Y+ +D
Sbjct 2 ATTYGTTSGAA----SANYDMSLWYDSKYYKIGMLTMLLVAIFWIWYQRTFAYSHGMDSM 57
Query 253 QPEFDRI 259
+PEFD++
Sbjct 58 EPEFDKV 64
> nit:NAL212_1386 methane monooxygenase/ammonia monooxygenase
subunit C
Length=269
Score = 39.3 bits (90), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 21/67 (31%), Positives = 37/67 (55%), Gaps = 4/67 (6%)
Query 193 AEALGSKSGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHY 252
A G+ SG A S + ++ ++ D + Y IG+LT+ V I+ + F Y+ +D
Sbjct 2 ATTYGTTSGAA----SANYDMSLWYDSKYYKIGMLTMLLVAIFWIWYQRTFAYSHGMDSM 57
Query 253 QPEFDRI 259
+PEFD++
Sbjct 58 EPEFDKV 64
> nit:NAL212_0797 methane monooxygenase/ammonia monooxygenase
subunit C
Length=269
Score = 39.3 bits (90), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 21/67 (31%), Positives = 37/67 (55%), Gaps = 4/67 (6%)
Query 193 AEALGSKSGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHY 252
A G+ SG A S + ++ ++ D + Y IG+LT+ V I+ + F Y+ +D
Sbjct 2 ATTYGTTSGAA----SANYDMSLWYDSKYYKIGMLTMLLVAIFWIWYQRTFAYSHGMDSM 57
Query 253 QPEFDRI 259
+PEFD++
Sbjct 58 EPEFDKV 64
> nit:NAL212_2818 methane monooxygenase/ammonia monooxygenase
subunit C
Length=295
Score = 38.9 bits (89), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 24/92 (26%), Positives = 47/92 (51%), Gaps = 12/92 (13%)
Query 168 ISRELSMRTVEQTVDQQSTSSQGQYAEALGSKSGIAGVYGSTSNNIEVFCDEESYIIGLL 227
I+ L + ++Q D +T+ G+ SG A S + ++ ++ D + Y +G+L
Sbjct 11 INNYLRIFLIKQMEDMMATTY--------GTTSGAA----SANYDMSLWYDSKYYKLGML 58
Query 228 TVTPVPIYTQMLSKDFLYNGLLDHYQPEFDRI 259
T+ V I+ + F Y+ +D +PEFD++
Sbjct 59 TMLLVAIFWIWYQRTFAYSHGMDSMEPEFDKV 90
> nit:NAL212_2303 methane monooxygenase/ammonia monooxygenase
subunit C
Length=269
Score = 38.1 bits (87), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 20/67 (30%), Positives = 37/67 (55%), Gaps = 4/67 (6%)
Query 193 AEALGSKSGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHY 252
A G+ SG A S + ++ ++ D + Y +G+LT+ V I+ + F Y+ +D
Sbjct 2 ATTYGTTSGAA----SANYDMSLWYDSKYYKLGMLTMLLVAIFWIWYQRTFAYSHGMDSM 57
Query 253 QPEFDRI 259
+PEFD++
Sbjct 58 EPEFDKV 64
> nii:Nit79A3_2886 methane monooxygenase/ammonia monooxygenase
subunit C
Length=269
Score = 38.1 bits (87), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 36/64 (56%), Gaps = 6/64 (9%)
Query 202 IAGVYGSTSN------NIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPE 255
+A YG+TS ++ ++ D + Y IG+LT+ V I+ + F Y+ +D +PE
Sbjct 1 MATTYGTTSASSSANYDMSLWYDSKYYKIGMLTMLLVAIFWIWYQRTFAYSHGMDSMEPE 60
Query 256 FDRI 259
FD++
Sbjct 61 FDKV 64
> nii:Nit79A3_1595 methane monooxygenase/ammonia monooxygenase
subunit C
Length=269
Score = 38.1 bits (87), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 36/64 (56%), Gaps = 6/64 (9%)
Query 202 IAGVYGSTSN------NIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPE 255
+A YG+TS ++ ++ D + Y IG+LT+ V I+ + F Y+ +D +PE
Sbjct 1 MATTYGTTSASSSANYDMSLWYDSKYYKIGMLTMLLVAIFWIWYQRTFAYSHGMDSMEPE 60
Query 256 FDRI 259
FD++
Sbjct 61 FDKV 64
> nii:Nit79A3_1079 methane monooxygenase/ammonia monooxygenase
subunit C
Length=269
Score = 38.1 bits (87), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 36/64 (56%), Gaps = 6/64 (9%)
Query 202 IAGVYGSTSN------NIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPE 255
+A YG+TS ++ ++ D + Y IG+LT+ V I+ + F Y+ +D +PE
Sbjct 1 MATTYGTTSASSSANYDMSLWYDSKYYKIGMLTMLLVAIFWIWYQRTFAYSHGMDSMEPE 60
Query 256 FDRI 259
FD++
Sbjct 61 FDKV 64
> nii:Nit79A3_0471 methane monooxygenase/ammonia monooxygenase
subunit C
Length=269
Score = 38.1 bits (87), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 36/64 (56%), Gaps = 6/64 (9%)
Query 202 IAGVYGSTSN------NIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNGLLDHYQPE 255
+A YG+TS ++ ++ D + Y IG+LT+ V I+ + F Y+ +D +PE
Sbjct 1 MATTYGTTSASSSANYDMSLWYDSKYYKIGMLTMLLVAIFWIWYQRTFAYSHGMDSMEPE 60
Query 256 FDRI 259
FD++
Sbjct 61 FDKV 64
> bfo:BRAFLDRAFT_106126 hypothetical protein
Length=499
Score = 37.0 bits (84), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 22/63 (35%), Positives = 31/63 (49%), Gaps = 5/63 (8%)
Query 49 ADDGTYSIQKQTVLVDEDGS-----KYGISYKVSEDGESLVGVDYDPVSEKTPVTAINSY 103
ADDG + K V ++DGS G+ Y DG++ V D+ V EKT + S+
Sbjct 402 ADDGVTCVCKDLVFHNDDGSVQCVTACGLGYAPGSDGKTCVDGDHAEVGEKTTKILLCSF 461
Query 104 AEL 106
AE
Sbjct 462 AEF 464
> lro:LOCK900_2893 Hypothetical protein
Length=2619
Score = 37.0 bits (84), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 29/105 (28%), Positives = 46/105 (44%), Gaps = 9/105 (9%)
Query 151 DIDIRFDELLMPEFIGGISRELSMRTVEQTVDQQSTS--------SQGQYAEALGSKSGI 202
D+ +D GI + + E+TVD S + GQY+ L S +G
Sbjct 1961 DVSFEYDGKTKASEAKGIQVIVKLGETEKTVDLTSADIVVANDDVNAGQYSYQL-SDAGK 2019
Query 203 AGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNG 247
A + +T NN ++ D+ + I G +T+TP + F YNG
Sbjct 2020 AKLQAATGNNYQLTADDLAKITGTITITPAVATADSNNVSFEYNG 2064
> lrl:LC705_02890 adhesion exoprotein
Length=2299
Score = 37.0 bits (84), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 26/105 (25%), Positives = 47/105 (45%), Gaps = 9/105 (9%)
Query 151 DIDIRFDELLMPEFIGGISRELSMRTVEQTVDQQSTS--------SQGQYAEALGSKSGI 202
D+ +D GI +++ E+TVD S G+Y+ L S +G
Sbjct 873 DVSFEYDGKTKASEAKGIQATVTLGETEKTVDLTSADIVVENDGVDAGKYSYQL-SDAGK 931
Query 203 AGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNG 247
A + +T NN ++ D+ + ++G +T+TP + F Y+G
Sbjct 932 AKLQAATGNNYQLTADDLAKVMGTITITPAAVTADSNDVSFEYDG 976
> lrc:LOCK908_2979 Adhesion exoprotein
Length=2299
Score = 37.0 bits (84), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 26/105 (25%), Positives = 47/105 (45%), Gaps = 9/105 (9%)
Query 151 DIDIRFDELLMPEFIGGISRELSMRTVEQTVDQQSTS--------SQGQYAEALGSKSGI 202
D+ +D GI +++ E+TVD S G+Y+ L S +G
Sbjct 873 DVSFEYDGKTKASEAKGIQATVTLGETEKTVDLTSADIVVENDGVDAGKYSYQL-SDAGK 931
Query 203 AGVYGSTSNNIEVFCDEESYIIGLLTVTPVPIYTQMLSKDFLYNG 247
A + +T NN ++ D+ + ++G +T+TP + F Y+G
Sbjct 932 AKLQAATGNNYQLTADDLAKVMGTITITPAAVTADSNDVSFEYDG 976
> cmk:103186980 cops2; COP9 signalosome subunit 2
Length=472
Score = 36.6 bits (83), Expect = 9.0, Method: Compositional matrix adjust.
Identities = 39/148 (26%), Positives = 69/148 (47%), Gaps = 29/148 (20%)
Query 65 EDGSKYGISYKVSEDGESLVGVDYD-------PVSEKTPVTAINSYAELAALTTTEGS-G 116
+D Y + Y SED S VD + + E P A++S+ ++ L +G G
Sbjct 11 DDEEDYDLEY--SEDSNSEPNVDLENQYYNSKALKEDDPKAALSSFQKVLELEGEKGEWG 68
Query 117 FTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFI-GGISRELSMR 175
F A ++ +++N + G +Y ++M R+ +LL +I ++R S +
Sbjct 69 FK--------ALKQMIKINFKLG-NYPEMMN-------RYKQLLT--YIRSAVTRNYSEK 110
Query 176 TVEQTVDQQSTSSQGQYAEALGSKSGIA 203
++ +D STS Q Q ++ GS GIA
Sbjct 111 SINSILDYISTSKQQQKSKRQGSSGGIA 138
Lambda K H a alpha
0.317 0.135 0.394 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 725577540634