bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-37_CDS_annotation_glimmer3.pl_2_2
Length=179
Score E
Sequences producing significant alignments: (Bits) Value
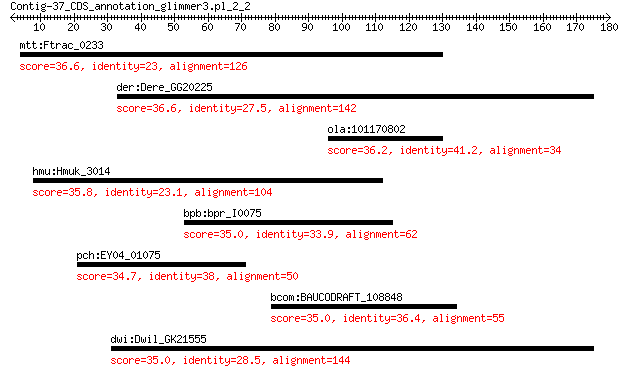
mtt:Ftrac_0233 restriction modification system DNA specificity... 36.6 2.2
der:Dere_GG20225 GG20225 gene product from transcript GG20225-RA 36.6 3.1
ola:101170802 thyroid receptor-interacting protein 11-like 36.2 3.8
hmu:Hmuk_3014 hypothetical protein 35.8 3.8
bpb:bpr_I0075 hypothetical protein 35.0 6.3
pch:EY04_01075 integrase 34.7 8.1
bcom:BAUCODRAFT_108848 glycosyltransferase family 2 protein 35.0 9.3
dwi:Dwil_GK21555 GK21555 gene product from transcript GK21555-RA 35.0 9.5
> mtt:Ftrac_0233 restriction modification system DNA specificity
domain
Length=505
Score = 36.6 bits (83), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 29/126 (23%), Positives = 58/126 (46%), Gaps = 19/126 (15%)
Query 4 KAQEIMNKYLPAQQQMQLYSYSANLFSQYAQGLLSLATIKNRLAEYNESLARTRGLNISN 63
K+Q+I + P +Q L S LFS+ G+ +L + K +L Y +++
Sbjct 131 KSQKI--PFAPLPEQKSLVSKIEQLFSELDNGIANLKSAKEKLEVYRQAV---------- 178
Query 64 EQSEKLSEYFIRAMKEEYQANAAYYRSYKSIAGAVATGRGNLDIYDSQLSRLQRDMQELF 123
L + F + +E++ S + ++ + GR L Y++Q+ Q+D+ E
Sbjct 179 -----LKKAFEGELTKEWRKKQTELPSAEDLSNQITFGRNKL--YENQIKEWQQDLVEWN 231
Query 124 ASRERY 129
+ R++Y
Sbjct 232 SKRKQY 237
> der:Dere_GG20225 GG20225 gene product from transcript GG20225-RA
Length=2347
Score = 36.6 bits (83), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 39/161 (24%), Positives = 66/161 (41%), Gaps = 24/161 (15%)
Query 33 AQGLLSLATIKNRLAEYNESLARTRGLNISNEQSEKLSEYFIRAMKEEYQANAAYYRSYK 92
A+ + L +++ L E N L R L +E+ K + + + E ANA + YK
Sbjct 1494 AESVAELEKVRSELEEVNNQL---RALKDEHEKITKECDELKKRTEPETDANA-VRQDYK 1549
Query 93 S---------------IAGAVATGRGNLDIYDSQLSRLQRDMQELFAS----RERYSMMN 133
+ +A T G YD + RL++++QE A+ +R + N
Sbjct 1550 AKLDKLVVDLTVARTELANQETTFAGTKSSYDETIGRLEKELQENIAANKDINQRLTREN 1609
Query 134 RKAYYRAQQIWRDFIGGIMSGAAAGTGVGAMTNFSRSSHKT 174
+ R Q+ R +G S + + V N S SS +T
Sbjct 1610 ESLHMRINQLTRQ-LGSQQSTKPSTSSVAEKGNISESSPRT 1649
> ola:101170802 thyroid receptor-interacting protein 11-like
Length=1599
Score = 36.2 bits (82), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 14/34 (41%), Positives = 24/34 (71%), Gaps = 0/34 (0%)
Query 96 GAVATGRGNLDIYDSQLSRLQRDMQELFASRERY 129
G V RG LD+ +++L+ L+R++ EL +SRE+
Sbjct 319 GEVERARGELDVKETELTELRRELDELRSSREKV 352
> hmu:Hmuk_3014 hypothetical protein
Length=447
Score = 35.8 bits (81), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 24/104 (23%), Positives = 55/104 (53%), Gaps = 0/104 (0%)
Query 8 IMNKYLPAQQQMQLYSYSANLFSQYAQGLLSLATIKNRLAEYNESLARTRGLNISNEQSE 67
++ +Y+ A ++M ++ + +Q + L+T + + Y+++ AR RG++ ++ SE
Sbjct 247 VLFRYIAAVERMTDENFELQTITSGSQEVARLSTNGDDIVLYHDNSARDRGVSFESDVSE 306
Query 68 KLSEYFIRAMKEEYQANAAYYRSYKSIAGAVATGRGNLDIYDSQ 111
K S+ R + E +A A +++ ATGR ++ + + Q
Sbjct 307 KPSKALSRTERIEREARAVADSYFRNRTFRSATGRPDVIVLEVQ 350
> bpb:bpr_I0075 hypothetical protein
Length=360
Score = 35.0 bits (79), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 21/67 (31%), Positives = 38/67 (57%), Gaps = 8/67 (12%)
Query 53 LARTRGLNISNEQSEKLSEY-----FIRAMKEEYQANAAYYRSYKSIAGAVATGRGNLDI 107
+A +G++++ ++SE+LSEY ++R KE+ Q A+ +R G T GN+D
Sbjct 203 VAVAKGIDLTPQESEQLSEYASDDNYVRTQKEDLQELASIFRDK---MGQDDTAVGNVDE 259
Query 108 YDSQLSR 114
Y L++
Sbjct 260 YVKLLNK 266
> pch:EY04_01075 integrase
Length=326
Score = 34.7 bits (78), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 19/50 (38%), Positives = 30/50 (60%), Gaps = 5/50 (10%)
Query 21 LYSYSANLFSQYAQGLLSLATIKNRLAEYNESLARTRGLNISNEQSEKLS 70
L Y+ +L +Q QG L++AT +NRL+ N ++A RG +QS K+
Sbjct 71 LLDYAGHLRNQVEQGSLAIATAQNRLSSVNRTMAALRG-----DQSVKVP 115
> bcom:BAUCODRAFT_108848 glycosyltransferase family 2 protein
Length=1853
Score = 35.0 bits (79), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 20/56 (36%), Positives = 31/56 (55%), Gaps = 1/56 (2%)
Query 79 EEYQANA-AYYRSYKSIAGAVATGRGNLDIYDSQLSRLQRDMQELFASRERYSMMN 133
EE Q A +Y+ + +++ G + G G L I D Q+ R + DMQ L + R R+ N
Sbjct 437 EEVQVPATSYFDNSEAVKGLLKPGNGLLSILDDQMRRGKSDMQFLESIRRRFEGKN 492
> dwi:Dwil_GK21555 GK21555 gene product from transcript GK21555-RA
Length=2361
Score = 35.0 bits (79), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 41/167 (25%), Positives = 70/167 (42%), Gaps = 30/167 (18%)
Query 31 QYAQGLLSLATIKNRLAEYNESLARTRGLNISNEQSEKLS----EYFIRAMKEEYQANAA 86
Q A + L +++ L E N L + E EKL+ E R + ANAA
Sbjct 1499 QTADTISELEKVRSELDEVNGQLRACK------EDQEKLTQANEELKKRVEPQTDAANAA 1552
Query 87 YYRSYKS---------------IAGAVATGRGNLDIYDSQLSRLQRDMQE-LFASRE--- 127
+ YK+ +A + G +YD ++RL++++Q+ + A++E
Sbjct 1553 VRQEYKAKLDKLVLDLTVARTDLANQETSFAGTRTVYDESIARLEKELQDTITANKEVNL 1612
Query 128 RYSMMNRKAYYRAQQIWRDFIGGIMSGAAAGTGVGAMTNFSRSSHKT 174
R + N + R Q+ R +G S + + V N S SS +T
Sbjct 1613 RLTRENEALHMRVNQLTRQ-LGTQQSTKPSTSSVAEKGNISDSSPRT 1658
Lambda K H a alpha
0.317 0.128 0.350 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 160264878837