bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-37_CDS_annotation_glimmer3.pl_2_3
Length=91
Score E
Sequences producing significant alignments: (Bits) Value
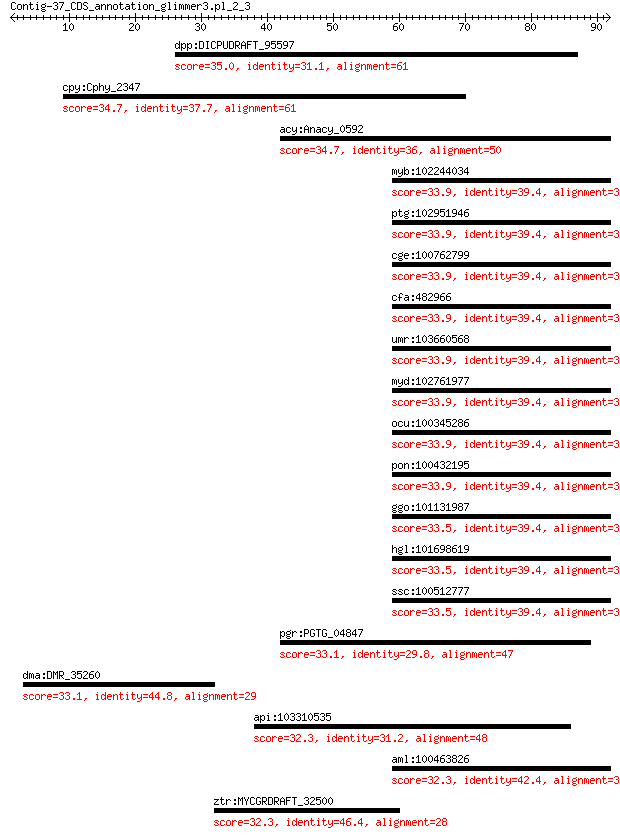
dpp:DICPUDRAFT_95597 hypothetical protein 35.0 1.3
cpy:Cphy_2347 extracellular solute-binding protein 34.7 1.6
acy:Anacy_0592 Tetratricopeptide TPR_1 repeat-containing protein 34.7 1.8
myb:102244034 NOL10; nucleolar protein 10 33.9
ptg:102951946 NOL10; nucleolar protein 10 33.9
cge:100762799 Nol10; nucleolar protein 10 33.9
cfa:482966 NOL10; nucleolar protein 10 33.9
umr:103660568 NOL10; nucleolar protein 10 33.9
myd:102761977 NOL10; nucleolar protein 10 33.9
ocu:100345286 NOL10; nucleolar protein 10 33.9
pon:100432195 NOL10; nucleolar protein 10 33.9
ggo:101131987 nucleolar protein 10-like 33.5 3.7
hgl:101698619 Nol10; nucleolar protein 10 33.5
ssc:100512777 NOL10; nucleolar protein 10 33.5
pgr:PGTG_04847 hypothetical protein 33.1 5.8
dma:DMR_35260 hypothetical protein 33.1 5.8
api:103310535 uncharacterized LOC103310535 32.3 8.2
aml:100463826 NOL10; nucleolar protein 10 32.3
ztr:MYCGRDRAFT_32500 hypothetical protein 32.3 9.5
> dpp:DICPUDRAFT_95597 hypothetical protein
Length=457
Score = 35.0 bits (79), Expect = 1.3, Method: Composition-based stats.
Identities = 19/64 (30%), Positives = 32/64 (50%), Gaps = 5/64 (8%)
Query 26 VFTMLLDGTLLPVYDFSGNNVRSSLNSLRK---YYPDSVVKFHFESSFYTQNTLPCPYHF 82
+F LL G L ++ N ++ N+L+K Y PD + K+H ++ Y T+ F
Sbjct 384 IFQQLLKGLGLDSFEIIPNELKEIYNTLKKTEFYDPDEICKYHARAALYDLETI--TKQF 441
Query 83 HYPD 86
+PD
Sbjct 442 IFPD 445
> cpy:Cphy_2347 extracellular solute-binding protein
Length=431
Score = 34.7 bits (78), Expect = 1.6, Method: Composition-based stats.
Identities = 23/62 (37%), Positives = 34/62 (55%), Gaps = 6/62 (10%)
Query 9 VNVEKAN-AIYSVFKTCNVFTMLLDGTLLPVYDFSGNNVRSSLNSLRKYYPDSVVKFHFE 67
VN K N AIY++ T N F M D +LL + N+++S + K P +V F+FE
Sbjct 148 VNTVKVNDAIYAIPFTHNTFFMYYDKSLL-----NENDIKSIEGIMAKETPSNVYNFYFE 202
Query 68 SS 69
S+
Sbjct 203 SA 204
> acy:Anacy_0592 Tetratricopeptide TPR_1 repeat-containing protein
Length=845
Score = 34.7 bits (78), Expect = 1.8, Method: Composition-based stats.
Identities = 18/53 (34%), Positives = 27/53 (51%), Gaps = 3/53 (6%)
Query 42 SGNNVRSSLNSLRKYYPDSVVKFHFESSFYTQNTLPCPYH--FHY-PDCILFF 91
+G+ +++ YYP+S ES+ +LPCPY FH+ PD FF
Sbjct 21 TGDRNTATITITNYYYPESTKVLPVESTGAADKSLPCPYRGLFHFGPDDAEFF 73
> myb:102244034 NOL10; nucleolar protein 10
Length=680
Score = 33.9 bits (76), Expect = 2.8, Method: Composition-based stats.
Identities = 13/36 (36%), Positives = 21/36 (58%), Gaps = 3/36 (8%)
Query 59 DSVVKFHFESSFYTQNTLPC---PYHFHYPDCILFF 91
D ++FH +S FY + +P + +HYP C L+F
Sbjct 108 DRYIEFHSQSGFYYKTRIPKFGRDFSYHYPSCDLYF 143
> ptg:102951946 NOL10; nucleolar protein 10
Length=736
Score = 33.9 bits (76), Expect = 2.9, Method: Composition-based stats.
Identities = 13/36 (36%), Positives = 21/36 (58%), Gaps = 3/36 (8%)
Query 59 DSVVKFHFESSFYTQNTLPC---PYHFHYPDCILFF 91
D ++FH +S FY + +P + +HYP C L+F
Sbjct 164 DRYIEFHSQSGFYYKTRIPKFGRDFSYHYPSCDLYF 199
> cge:100762799 Nol10; nucleolar protein 10
Length=690
Score = 33.9 bits (76), Expect = 3.3, Method: Composition-based stats.
Identities = 13/36 (36%), Positives = 21/36 (58%), Gaps = 3/36 (8%)
Query 59 DSVVKFHFESSFYTQNTLPC---PYHFHYPDCILFF 91
D ++FH +S FY + +P + +HYP C L+F
Sbjct 116 DRYIEFHSQSGFYYKTRIPKFGRDFSYHYPSCDLYF 151
> cfa:482966 NOL10; nucleolar protein 10
Length=662
Score = 33.9 bits (76), Expect = 3.4, Method: Composition-based stats.
Identities = 13/36 (36%), Positives = 21/36 (58%), Gaps = 3/36 (8%)
Query 59 DSVVKFHFESSFYTQNTLPC---PYHFHYPDCILFF 91
D ++FH +S FY + +P + +HYP C L+F
Sbjct 90 DRYIEFHSQSGFYYKTRIPKFGRDFSYHYPSCDLYF 125
> umr:103660568 NOL10; nucleolar protein 10
Length=686
Score = 33.9 bits (76), Expect = 3.4, Method: Composition-based stats.
Identities = 13/36 (36%), Positives = 21/36 (58%), Gaps = 3/36 (8%)
Query 59 DSVVKFHFESSFYTQNTLPC---PYHFHYPDCILFF 91
D ++FH +S FY + +P + +HYP C L+F
Sbjct 116 DRYIEFHSQSGFYYKTRIPKFGRDFSYHYPSCDLYF 151
> myd:102761977 NOL10; nucleolar protein 10
Length=688
Score = 33.9 bits (76), Expect = 3.4, Method: Composition-based stats.
Identities = 13/36 (36%), Positives = 21/36 (58%), Gaps = 3/36 (8%)
Query 59 DSVVKFHFESSFYTQNTLPC---PYHFHYPDCILFF 91
D ++FH +S FY + +P + +HYP C L+F
Sbjct 116 DRYIEFHSQSGFYYKTRIPKFGRDFSYHYPSCDLYF 151
> ocu:100345286 NOL10; nucleolar protein 10
Length=688
Score = 33.9 bits (76), Expect = 3.4, Method: Composition-based stats.
Identities = 13/36 (36%), Positives = 21/36 (58%), Gaps = 3/36 (8%)
Query 59 DSVVKFHFESSFYTQNTLPC---PYHFHYPDCILFF 91
D ++FH +S FY + +P + +HYP C L+F
Sbjct 116 DRYIEFHSQSGFYYKTRIPKFGRDFSYHYPSCDLYF 151
> pon:100432195 NOL10; nucleolar protein 10
Length=688
Score = 33.9 bits (76), Expect = 3.4, Method: Composition-based stats.
Identities = 13/36 (36%), Positives = 21/36 (58%), Gaps = 3/36 (8%)
Query 59 DSVVKFHFESSFYTQNTLPC---PYHFHYPDCILFF 91
D ++FH +S FY + +P + +HYP C L+F
Sbjct 116 DRYIEFHSQSGFYYKTRIPKFGRDFSYHYPSCDLYF 151
> ggo:101131987 nucleolar protein 10-like
Length=399
Score = 33.5 bits (75), Expect = 3.7, Method: Composition-based stats.
Identities = 13/36 (36%), Positives = 21/36 (58%), Gaps = 3/36 (8%)
Query 59 DSVVKFHFESSFYTQNTLPC---PYHFHYPDCILFF 91
D ++FH +S FY + +P + +HYP C L+F
Sbjct 97 DRYIEFHSQSGFYYKTRIPKFGRDFSYHYPSCDLYF 132
> hgl:101698619 Nol10; nucleolar protein 10
Length=688
Score = 33.5 bits (75), Expect = 3.7, Method: Composition-based stats.
Identities = 13/36 (36%), Positives = 21/36 (58%), Gaps = 3/36 (8%)
Query 59 DSVVKFHFESSFYTQNTLPC---PYHFHYPDCILFF 91
D ++FH +S FY + +P + +HYP C L+F
Sbjct 116 DRYIEFHSQSGFYYKTRIPKFGRDFSYHYPSCDLYF 151
> ssc:100512777 NOL10; nucleolar protein 10
Length=678
Score = 33.5 bits (75), Expect = 4.0, Method: Composition-based stats.
Identities = 13/36 (36%), Positives = 21/36 (58%), Gaps = 3/36 (8%)
Query 59 DSVVKFHFESSFYTQNTLPC---PYHFHYPDCILFF 91
D ++FH +S FY + +P + +HYP C L+F
Sbjct 116 DRYIEFHSQSGFYYKTRIPKFGRDFSYHYPSCDLYF 151
> pgr:PGTG_04847 hypothetical protein
Length=610
Score = 33.1 bits (74), Expect = 5.8, Method: Composition-based stats.
Identities = 14/47 (30%), Positives = 25/47 (53%), Gaps = 3/47 (6%)
Query 42 SGNNVRSSLNSLRKYYPDSVVKFHFESSFYTQNTLPCPYHFHYPDCI 88
S + R++L ++K P V +H E Y + P+ +H+PDC+
Sbjct 121 SPEDFRTALEKIQKILPVDCVSWHREDLLYHGTS---PWTYHHPDCL 164
> dma:DMR_35260 hypothetical protein
Length=1062
Score = 33.1 bits (74), Expect = 5.8, Method: Composition-based stats.
Identities = 13/29 (45%), Positives = 20/29 (69%), Gaps = 0/29 (0%)
Query 3 TNKLFRVNVEKANAIYSVFKTCNVFTMLL 31
T LF V++ +A +Y+ F C+VF+MLL
Sbjct 156 TGSLFHVDISRAIKLYATFFCCSVFSMLL 184
> api:103310535 uncharacterized LOC103310535
Length=219
Score = 32.3 bits (72), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 15/48 (31%), Positives = 25/48 (52%), Gaps = 6/48 (13%)
Query 38 VYDFSGNNVRSSLNSLRKYYPDSVVKFHFESSFYTQNTLPCPYHFHYP 85
+Y G N + + + LR+ + D V + F NT+PC +HF+ P
Sbjct 103 IYSNCGTNFQDAGSELRRLWSDPVAQNTFS------NTIPCRWHFNPP 144
> aml:100463826 NOL10; nucleolar protein 10
Length=688
Score = 32.3 bits (72), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 14/36 (39%), Positives = 21/36 (58%), Gaps = 3/36 (8%)
Query 59 DSVVKFHFESSFYTQNTLPC---PYHFHYPDCILFF 91
D ++FH +S FY + +P + +HYP C LFF
Sbjct 116 DRYIEFHSQSGFYYKTRIPKFGRDFSYHYPSCDLFF 151
> ztr:MYCGRDRAFT_32500 hypothetical protein
Length=388
Score = 32.3 bits (72), Expect = 9.5, Method: Composition-based stats.
Identities = 13/28 (46%), Positives = 18/28 (64%), Gaps = 0/28 (0%)
Query 32 DGTLLPVYDFSGNNVRSSLNSLRKYYPD 59
+GT LP + GN V+++LNS YPD
Sbjct 218 NGTFLPFFQLGGNFVQANLNSALSKYPD 245
Lambda K H a alpha
0.327 0.140 0.443 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 127599116940