bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-3_CDS_annotation_glimmer3.pl_2_6
Length=369
Score E
Sequences producing significant alignments: (Bits) Value
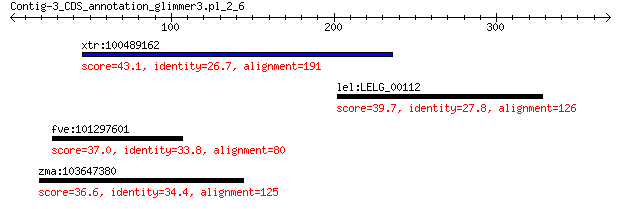
xtr:100489162 lamc2; laminin, gamma 2 43.1
lel:LELG_00112 hypothetical protein 39.7 1.2
fve:101297601 minor spike protein H-like 37.0 1.7
zma:103647380 uncharacterized LOC103647380 36.6 9.6
> xtr:100489162 lamc2; laminin, gamma 2
Length=1172
Score = 43.1 bits (100), Expect = 0.10, Method: Composition-based stats.
Identities = 51/192 (27%), Positives = 76/192 (40%), Gaps = 31/192 (16%)
Query 45 AEKARQFQLDMWNKTNEYNSASAQRSRLEEAGLNPYLMMNGGSAGTAQSSGSSTPASASS 104
+ K ++ + T E N S R LEE M A AQ + A+ +
Sbjct 612 STKLNLYKRQLLKLTGENNGQSPARGELEE-------RMRKAEAA-AQGLQRDSEAANGA 663
Query 105 PLSMQRQDFSGLSNTLASALQISNQTKETNANVQTLQSQKSLYDAQANSILSNVDWWKLG 164
LS+QRQ FSGL T + + +Q KE +Q LQ+QK+ Y Q + ++
Sbjct 664 ALSLQRQ-FSGLQGTQSDTQNVLDQAKE---KLQGLQTQKNQYQGQLQDMGRKINI---- 715
Query 165 PEYKKWSQMTGLARAGLQFQTDKQNLRNMTW-SGNLIQAQHIGALLDNKSKRIINNYLDE 223
A Q Q ++ L MT+ S NL + L +++ I N E
Sbjct 716 --------------ALQQLQGSQEELNRMTFPSTNLPLDSGSYSKLRQEAQEIANRLAQE 761
Query 224 GQRLQLDLMAAQ 235
Q + D AQ
Sbjct 762 AQTVAQDAAEAQ 773
> lel:LELG_00112 hypothetical protein
Length=660
Score = 39.7 bits (91), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 35/128 (27%), Positives = 64/128 (50%), Gaps = 8/128 (6%)
Query 202 AQHIGALLDNKSK-RIINNYLDEGQRLQLDLMAAQYYDAMASGHLKYQQAKSEITKRILM 260
AQ I A LD +K R++ N ++E +L+ ++A++ + LK + +++++R+L
Sbjct 474 AQRIEAQLDQVAKSRVVLNSINE----KLNTLSAKHDLGNTTRILKAKARHTQLSRRVLR 529
Query 261 MAEAKG-LQINNKAAEDTADGYIKALNAEYSASYDINSPFKYGEDSYVPASVLKSRMDAL 319
+A L++ +G K + S D +SP D + +VLK R + L
Sbjct 530 LATVLAILKLKGYPLLPEEEGIAKQFDLLSSKINDPSSPIGKLGDIFAKLTVLKERAEDL 589
Query 320 NSKWQFDK 327
NS QFD+
Sbjct 590 NS--QFDQ 595
> fve:101297601 minor spike protein H-like
Length=139
Score = 37.0 bits (84), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 27/82 (33%), Positives = 43/82 (52%), Gaps = 7/82 (9%)
Query 27 NNANMAVNRMNNEFNAAEAEKARQFQLDMWNKTNEYNSASAQRSRLEEAGLNPYL--MMN 84
N N A+ + E + AEA++ R+FQ + N + Q + L AGLNP L +
Sbjct 8 NATNEAIANQSTETSMAEAQRNREFQERLSNSAYQR-----QVADLSSAGLNPMLAYIKG 62
Query 85 GGSAGTAQSSGSSTPASASSPL 106
GG++ + S+G T A +SP+
Sbjct 63 GGASTPSGSTGQVTSAQYTSPI 84
> zma:103647380 uncharacterized LOC103647380
Length=1301
Score = 36.6 bits (83), Expect = 9.6, Method: Composition-based stats.
Identities = 43/137 (31%), Positives = 66/137 (48%), Gaps = 19/137 (14%)
Query 19 NVVSTNKANNANMAVNRMNNEFNAAEAEKARQFQLDMWNKTNEYNSASAQRSRLEEAGLN 78
NVVS KA N++ +MN +F+A + E+ Q +D K NE S S QRS L N
Sbjct 476 NVVS--KATQRNVSDEKMNGKFSAHKGEQVAQEMVDHKMK-NELES-SLQRSNLRTPLSN 531
Query 79 PYLM---------MNGGSAGTAQSSGSSTPASASSPLSMQRQDFSGLS---NTLASALQI 126
P + MN G G A+SS S + LS +++ S ++ N A L+
Sbjct 532 PIDVKVKRLDSGKMNAGLLGNARSSAGGKMQSKT--LSAKKEVSSNMNTKQNKFAHKLK- 588
Query 127 SNQTKETNANVQTLQSQ 143
S+ T + N ++ +S+
Sbjct 589 SDDTSKGNLHLSARESK 605
Lambda K H a alpha
0.311 0.125 0.351 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 710007207144