bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-40_CDS_annotation_glimmer3.pl_2_1
Length=297
Score E
Sequences producing significant alignments: (Bits) Value
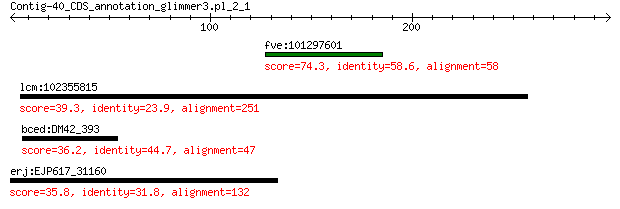
fve:101297601 minor spike protein H-like 74.3 1e-13
lcm:102355815 SCYL2; SCY1-like 2 (S. cerevisiae) 39.3 0.99
bced:DM42_393 hypothetical protein 36.2 8.2
erj:EJP617_31160 hypothetical protein 35.8 8.5
> fve:101297601 minor spike protein H-like
Length=139
Score = 74.3 bits (181), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 34/59 (58%), Positives = 45/59 (76%), Gaps = 1/59 (2%)
Query 127 EAMKNREWQEHMSNTAYQRAVADMKEAGLNPILAF-QNGGASTPGGSAGTISGASIGAP 184
EA +NRE+QE +SN+AYQR VAD+ AGLNP+LA+ + GGASTP GS G ++ A +P
Sbjct 25 EAQRNREFQERLSNSAYQRQVADLSSAGLNPMLAYIKGGGASTPSGSTGQVTSAQYTSP 83
> lcm:102355815 SCYL2; SCY1-like 2 (S. cerevisiae)
Length=936
Score = 39.3 bits (90), Expect = 0.99, Method: Composition-based stats.
Identities = 60/265 (23%), Positives = 98/265 (37%), Gaps = 49/265 (18%)
Query 6 LGALGTIGNVAGTVGTVANAVGALGNAFGAWGQVGQSQSQGGSTQQGGGSSQSMSKSGTN 65
L G +G + T N + + N FG Q G S + T G +Q +K
Sbjct 623 LDVTGQMGKLEETKPNTDNQIDKIFNNFGVSAQAGGSAGKENGTSAG---AQQPAKGSLT 679
Query 66 DEQVMQYLKGAYQYQNAEGQ--------------RQSQFNQRSMLEQMGYNTLGAIAQGI 111
E+ + K Q Q Q +Q++ S+L+ M LG ++
Sbjct 680 LEEKQKLAKEQEQAQKLRNQQPLMPKSVTPTAPVKQTKDLTGSLLDNMA--NLGNLSVST 737
Query 112 YNHIENSAAMNFNSTEAMKNREWQEHMSNTAYQRAVADMKEAGLNPILAFQNGGASTPGG 171
+ S + ++S M + M TA V M G NP + FQ+ G S G
Sbjct 738 TKPSQVSVSSGYSSAHPMGSS-----MGFTAPSSNVGTMSN-GFNPNMGFQSSGFSM--G 789
Query 172 SAGTISGASIGAPSSSALGVSRASGFVPNSYSSESWSQSDWYNAAQSWNQMLSSTGMTPL 231
+GT GA S G++ AS ++ ++N M + G+T
Sbjct 790 MSGTNQSFFSGASSVGMGGMTTASA-----------------SSMPNFNSMAGTVGITQQ 832
Query 232 GLQETLSDIGKNAGNAIDNAIGAGR 256
Q +D+ +A+DN G+ +
Sbjct 833 TQQSKATDM-----SALDNLFGSQK 852
> bced:DM42_393 hypothetical protein
Length=580
Score = 36.2 bits (82), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 21/47 (45%), Positives = 26/47 (55%), Gaps = 0/47 (0%)
Query 7 GALGTIGNVAGTVGTVANAVGALGNAFGAWGQVGQSQSQGGSTQQGG 53
G L + NVAG G VANA G G A GQ G++ GG+ +GG
Sbjct 375 GELADVANVAGKTGEVANAAGKTGEVAEAAGQAGKASDIGGAAAKGG 421
> erj:EJP617_31160 hypothetical protein
Length=346
Score = 35.8 bits (81), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 42/136 (31%), Positives = 71/136 (52%), Gaps = 11/136 (8%)
Query 1 MAFSLLG--ALGTIGNVAG-TVGTVANAVGALGNAFGAW-GQVGQSQSQGGSTQQGGGSS 56
+ ++LG A+G + N AG VG +A+A G++ + G+ GQ Q G G
Sbjct 99 LVATILGFAAVGGVLNTAGNAVGGIASATGSMASGIGSVAGQAAQGAGNMGEKIYGRLGL 158
Query 57 QSMSKSGTNDEQVMQYLKGAYQYQNAEGQRQSQFNQRSMLEQMGYNTLGAIAQGIYNHIE 116
+ +S D+QVM+ LK ++ + Q ++ Q S LE G + L + A+ + +H E
Sbjct 159 DTRLQSQDADQQVMEALK-----KSGIKELQPEYLQ-SQLEAAG-DDLASAAKNLASHPE 211
Query 117 NSAAMNFNSTEAMKNR 132
NS A+ + TE +K+R
Sbjct 212 NSDAIVNSLTEKLKSR 227
Lambda K H a alpha
0.308 0.124 0.353 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 501929306787