bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-6_CDS_annotation_glimmer3.pl_2_1
Length=153
Score E
Sequences producing significant alignments: (Bits) Value
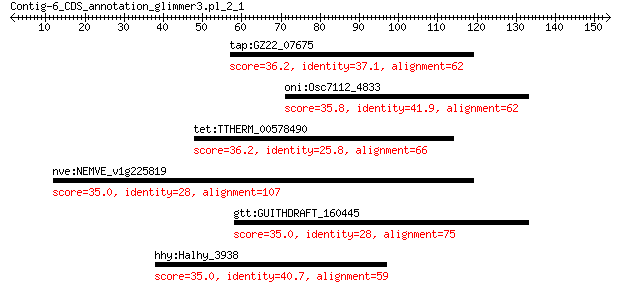
tap:GZ22_07675 TetR family transcriptional regulator 36.2 1.3
oni:Osc7112_4833 CHAD domain containing protein 35.8 2.1
tet:TTHERM_00578490 DHHC zinc finger domain containing protein 36.2 2.3
nve:NEMVE_v1g225819 hypothetical protein 35.0 3.8
gtt:GUITHDRAFT_160445 hypothetical protein 35.0 4.3
hhy:Halhy_3938 Fis family transcriptional regulator 35.0 5.0
> tap:GZ22_07675 TetR family transcriptional regulator
Length=183
Score = 36.2 bits (82), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 23/63 (37%), Positives = 34/63 (54%), Gaps = 3/63 (5%)
Query 57 ENKLTASELREYIQRYTPNKS-VYTSQLDDDTLLNTLKSRHIQSLSEMRSWAEYCMENYD 115
E K A +++ R N+S +Y LD LL+ L +HIQ+L+EM WA C E ++
Sbjct 26 EKKFEAITIQDISDRANLNRSTIYLHYLDKYDLLDKLIDQHIQTLTEMDIWA--CEEEWE 83
Query 116 SLI 118
I
Sbjct 84 DAI 86
> oni:Osc7112_4833 CHAD domain containing protein
Length=338
Score = 35.8 bits (81), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 26/65 (40%), Positives = 39/65 (60%), Gaps = 6/65 (9%)
Query 71 RYTPNKSVYTSQLDDDTLLNTLKSRHIQSLSEMRSWAEYCMENYDSL---IKDAEEKARV 127
RY PN S ++ D LLN LK RH ++L E+R+ E+ ++Y+SL I+D EK +
Sbjct 102 RYYPNLSADEQEVLDKALLNLLKQRH-RALKEVRAILEH--KSYESLKQSIEDWLEKPKF 158
Query 128 AAEEN 132
A E+
Sbjct 159 QAIEH 163
> tet:TTHERM_00578490 DHHC zinc finger domain containing protein
Length=1007
Score = 36.2 bits (82), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 17/67 (25%), Positives = 39/67 (58%), Gaps = 1/67 (1%)
Query 48 SLIFNQQRLENKLTASELREYIQRYTPNKSVYTSQLDDDTLLNTLKSRHIQSLSE-MRSW 106
SLIF ++ ++KL +L+EYI++Y PN + ++L + K + +++ +
Sbjct 784 SLIFEKEYYDSKLLMKQLKEYIEQYHPNNPLLQAELQYQEIHFLYKKKKFNEINDKINQA 843
Query 107 AEYCMEN 113
+++C++N
Sbjct 844 SQFCLQN 850
> nve:NEMVE_v1g225819 hypothetical protein
Length=265
Score = 35.0 bits (79), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 30/110 (27%), Positives = 55/110 (50%), Gaps = 11/110 (10%)
Query 12 NGSFDVLHTKVPVQDKLMQLSTFVNKDGSIAISSDISL-IFNQQRLENKLTASELREYIQ 70
NGS D+ H V V D ++ +++ + I++ + L FN ++ A E+ +
Sbjct 4 NGSIDIPH--VAVTDHQIRKRPLTDEEAA-EITAFLGLRCFNNDSVDGVTQARAFLEFYE 60
Query 71 RYTPNKSVYTSQLDDDTLLNTLKSRHIQSLSEMRSW--AEYCMENYDSLI 118
RY PNK + S + LN K++ +++ + R + A Y + NYD +I
Sbjct 61 RYNPNKGLLDSAIG---YLN--KAQKLEAQKQNRDYIRAYYLLNNYDKVI 105
> gtt:GUITHDRAFT_160445 hypothetical protein
Length=348
Score = 35.0 bits (79), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 21/75 (28%), Positives = 42/75 (56%), Gaps = 0/75 (0%)
Query 58 NKLTASELREYIQRYTPNKSVYTSQLDDDTLLNTLKSRHIQSLSEMRSWAEYCMENYDSL 117
++LTA+EL+ +QRY P +V+ S +L+ + + I+ + M+ + + YD++
Sbjct 273 DRLTANELQRIVQRYKPYLAVWFSSESAQEVLDVMSDQFIRMSNFMKRNFQIQFQRYDTV 332
Query 118 IKDAEEKARVAAEEN 132
K EE ++ + EE
Sbjct 333 KKLQEELSKTSKEEQ 347
> hhy:Halhy_3938 Fis family transcriptional regulator
Length=456
Score = 35.0 bits (79), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 24/68 (35%), Positives = 36/68 (53%), Gaps = 9/68 (13%)
Query 38 DGSIAISSDISLIFNQQRLENKLTAS--ELREYIQRYTP-------NKSVYTSQLDDDTL 88
D IAI + +SL+F QQ E + +S E ++Q++TP N S+ TS D +
Sbjct 6 DDDIAIQTSLSLLFQQQGFEVRTASSPTEALTFLQKHTPELAILDMNFSIETSGSDGLST 65
Query 89 LNTLKSRH 96
L +KS H
Sbjct 66 LRKIKSLH 73
Lambda K H a alpha
0.309 0.122 0.323 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 128392897951