bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-6_CDS_annotation_glimmer3.pl_2_6
Length=587
Score E
Sequences producing significant alignments: (Bits) Value
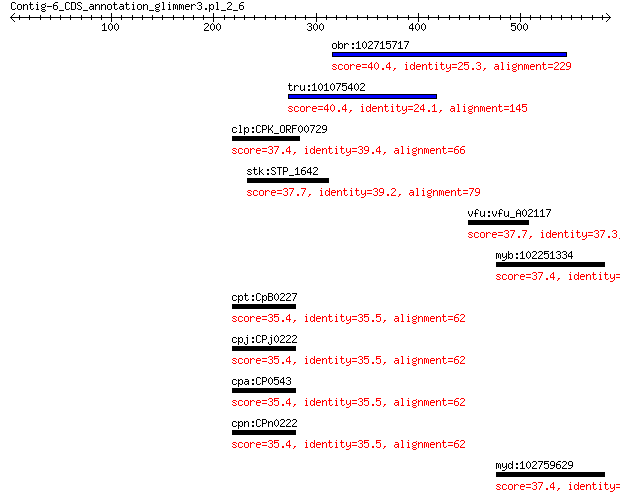
obr:102715717 uncharacterized LOC102715717 40.4 0.92
tru:101075402 endothelin-converting enzyme 1-like 40.4 1.3
clp:CPK_ORF00729 hypothetical protein 37.4 2.0
stk:STP_1642 hypothetical protein 37.7 3.5
vfu:vfu_A02117 hypothetical protein 37.7 5.6
myb:102251334 chromosome unknown open reading frame, human C20... 37.4 7.6
cpt:CpB0227 hypothetical protein 35.4 8.7
cpj:CPj0222 hypothetical protein 35.4 8.7
cpa:CP0543 hypothetical protein 35.4 8.7
cpn:CPn0222 hypothetical protein 35.4 8.7
myd:102759629 chromosome unknown open reading frame, human C20... 37.4 9.4
> obr:102715717 uncharacterized LOC102715717
Length=431
Score = 40.4 bits (93), Expect = 0.92, Method: Compositional matrix adjust.
Identities = 58/282 (21%), Positives = 114/282 (40%), Gaps = 54/282 (19%)
Query 316 VSGYLNSVVSLPGIFTDVSFTKNKSRFSKLFGYESFRQTIK--------TPEQAIERLSE 367
V+ LN+ + +P F S ++ S+F +F + F +T+K P +ER +
Sbjct 53 VASLLNATLVIPA-FHFNSVWRDHSKFGDIFDEDHFIETLKEHVRVVKELPVDVLERFNH 111
Query 368 RILFVRNGKPCEFTPPFSYISRLLP--------RFVPYSSNFVVETRTVLRSIRGVL--Q 417
I + N + ++ P Y+ ++LP R P+S+ + L+++R + Q
Sbjct 112 NISSIPNMRTKAYSSPNHYMQKVLPKLRELGVVRIAPFSNRLAQSVPSNLQALRCFVNYQ 171
Query 418 LFRRNEPFKKETPTNISEFVHTYTVTLYE-------------KYGYC-YD-TLPECLRVY 462
R EP + + V T+T + + C YD L E + +
Sbjct 172 ALRFAEPIRVLAENMVERMVKRSTLTGGKYVSVHLRFEEDMVAFSCCIYDGGLKEKIEME 231
Query 463 LSYTRSVKEIHYFTDKLKNKLCR---------PLYIYRIWESLGLSDDCIISL------- 506
+ RS K + ++ N PL + + +G + + +
Sbjct 232 NARERSWKGKFHRHGRVINPEANRRDGKCPLTPLEVGMMLRGMGFDNTTSLYVASGKIYN 291
Query 507 SDEYMSRCRSM----SLEKQLSIQQEMFEREGYSDELLSLFY 544
+++YM+ R M + + L++ +E+ E EG+S L +L Y
Sbjct 292 AEKYMAPLRQMFPLLATKDTLALPEELAEFEGHSSRLAALDY 333
> tru:101075402 endothelin-converting enzyme 1-like
Length=766
Score = 40.4 bits (93), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 35/151 (23%), Positives = 63/151 (42%), Gaps = 9/151 (6%)
Query 273 PHFHILFFFDSDEVAKNIRQAVFQSWKLGRVDTQLARDSAGSYVSGYLNSVVSLPGIFTD 332
P F + DS N+ Q S L D L + + Y+S YLN +V L G+
Sbjct 232 PFFSVFVSTDSKNSNSNVIQIDQSSLGLPSRDYYLNKTAHDKYLSAYLNFLVDL-GVLLG 290
Query 333 VSFTKNKSRFSKLFGYESFRQTIKTPEQAIERLSERILF--VRNGKPCEFTPPFSYISRL 390
S +++ + + +E+ I P++ +R E +++ ++ E P ++ L
Sbjct 291 GSAQSSRTMMAAIVDFETALANISVPQE--KRRDEELIYHKMKAKDLAELVPAVDWMPYL 348
Query 391 LPRFVPYSSN----FVVETRTVLRSIRGVLQ 417
F P N VV + L+ + +LQ
Sbjct 349 TAVFAPVVLNESEPVVVYAKEYLQEVSDLLQ 379
> clp:CPK_ORF00729 hypothetical protein
Length=121
Score = 37.4 bits (85), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 26/67 (39%), Positives = 34/67 (51%), Gaps = 10/67 (15%)
Query 218 DNNAFPQFKGLLKYINYRDYQLFAKRFRKYLFTRIGSYEKISSYVVSEYSPKTFRPHFHI 277
D+ PQ+ L+K QLF KR R RI + KI + EY K RPH+H+
Sbjct 60 DDKHLPQYGSLVKL----HLQLFLKRLRD----RISPH-KIRYFGCGEYGTKLQRPHYHL 110
Query 278 LFF-FDS 283
L F +DS
Sbjct 111 LIFNYDS 117
> stk:STP_1642 hypothetical protein
Length=180
Score = 37.7 bits (86), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 31/95 (33%), Positives = 46/95 (48%), Gaps = 19/95 (20%)
Query 233 NYRDYQLFAKRFRKYLFTRIGSY----EKISSYVVSEYSPKTFRPHFHILFFFDSDEVA- 287
N+ DY+L AK+ K + T I + EK+S Y + K R + + F +EV+
Sbjct 29 NHIDYKLIAKQTSKSVST-IKKWRLRIEKLSGYSFKKAKHKVSRYSTQVFYEFTDEEVSK 87
Query 288 -----------KNIRQAVFQSWKLGRVDTQLARDS 311
KN+ QA+ Q W G +DTQL +DS
Sbjct 88 FIEVNNLVNKTKNLDQAIKQVW--GDLDTQLEQDS 120
> vfu:vfu_A02117 hypothetical protein
Length=266
Score = 37.7 bits (86), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 22/59 (37%), Positives = 32/59 (54%), Gaps = 6/59 (10%)
Query 449 GYCYDTLPECLRVYLSYTRSVKEIHYFTDKLKNKLCRPLYIYRIWESLGLSDDCIISLS 507
G YDT E + TR+VK+I DKLKN P+ E+L +SD+ I+S++
Sbjct 177 GDIYDTTTESEAIRSDLTRAVKDIRESIDKLKNIKTLPV------EALSISDEIILSIN 229
> myb:102251334 chromosome unknown open reading frame, human C20orf96
Length=358
Score = 37.4 bits (85), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 26/108 (24%), Positives = 56/108 (52%), Gaps = 2/108 (2%)
Query 476 TDKLKNKLCRPLYIYR--IWESLGLSDDCIISLSDEYMSRCRSMSLEKQLSIQQEMFERE 533
+ K+K +L R L R + L + +I L+ + ++ + M L + + +++
Sbjct 88 SGKIKLRLMRSLLRSRRTALQELCKQEGFLIKLNHDLITTIKDMEDSSALKTRGMLQQQD 147
Query 534 GYSDELLSLFYINKPQKKVNNRYFQEWKDKNYYEVHYIRVKHKKLNDE 581
+ + SL Y NK + + R QEWK+K +++ Y++ + ++LND+
Sbjct 148 IFGTIINSLGYSNKKKLQQMTRELQEWKEKEEFKMSYLKQQVEQLNDK 195
> cpt:CpB0227 hypothetical protein
Length=113
Score = 35.4 bits (80), Expect = 8.7, Method: Compositional matrix adjust.
Identities = 22/62 (35%), Positives = 29/62 (47%), Gaps = 9/62 (15%)
Query 218 DNNAFPQFKGLLKYINYRDYQLFAKRFRKYLFTRIGSYEKISSYVVSEYSPKTFRPHFHI 277
D+ PQ+ L+K QLF KR RK + S KI + Y K RPH+H+
Sbjct 60 DDKHLPQYGSLVKL----HLQLFLKRLRKMI-----SPHKIRYFECGAYGTKLQRPHYHL 110
Query 278 LF 279
L
Sbjct 111 LL 112
> cpj:CPj0222 hypothetical protein
Length=113
Score = 35.4 bits (80), Expect = 8.7, Method: Compositional matrix adjust.
Identities = 22/62 (35%), Positives = 29/62 (47%), Gaps = 9/62 (15%)
Query 218 DNNAFPQFKGLLKYINYRDYQLFAKRFRKYLFTRIGSYEKISSYVVSEYSPKTFRPHFHI 277
D+ PQ+ L+K QLF KR RK + S KI + Y K RPH+H+
Sbjct 60 DDKHLPQYGSLVKL----HLQLFLKRLRKMI-----SPHKIRYFECGAYGTKLQRPHYHL 110
Query 278 LF 279
L
Sbjct 111 LL 112
> cpa:CP0543 hypothetical protein
Length=113
Score = 35.4 bits (80), Expect = 8.7, Method: Compositional matrix adjust.
Identities = 22/62 (35%), Positives = 29/62 (47%), Gaps = 9/62 (15%)
Query 218 DNNAFPQFKGLLKYINYRDYQLFAKRFRKYLFTRIGSYEKISSYVVSEYSPKTFRPHFHI 277
D+ PQ+ L+K QLF KR RK + S KI + Y K RPH+H+
Sbjct 60 DDKHLPQYGSLVKL----HLQLFLKRLRKMI-----SPHKIRYFECGAYGTKLQRPHYHL 110
Query 278 LF 279
L
Sbjct 111 LL 112
> cpn:CPn0222 hypothetical protein
Length=113
Score = 35.4 bits (80), Expect = 8.7, Method: Compositional matrix adjust.
Identities = 22/62 (35%), Positives = 29/62 (47%), Gaps = 9/62 (15%)
Query 218 DNNAFPQFKGLLKYINYRDYQLFAKRFRKYLFTRIGSYEKISSYVVSEYSPKTFRPHFHI 277
D+ PQ+ L+K QLF KR RK + S KI + Y K RPH+H+
Sbjct 60 DDKHLPQYGSLVKL----HLQLFLKRLRKMI-----SPHKIRYFECGAYGTKLQRPHYHL 110
Query 278 LF 279
L
Sbjct 111 LL 112
> myd:102759629 chromosome unknown open reading frame, human C20orf96
Length=358
Score = 37.4 bits (85), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 27/108 (25%), Positives = 55/108 (51%), Gaps = 2/108 (2%)
Query 476 TDKLKNKLCRPLYIYR--IWESLGLSDDCIISLSDEYMSRCRSMSLEKQLSIQQEMFERE 533
+ K+K +L R L R + L + +I LS + ++ + M L + + +++
Sbjct 88 SGKIKLRLMRALLRSRRTALQELCKQEGFLIKLSQDLITTIKDMEDSSALKTRAMLQQQD 147
Query 534 GYSDELLSLFYINKPQKKVNNRYFQEWKDKNYYEVHYIRVKHKKLNDE 581
+ + SL Y NK + + R QEWK+K ++ Y+R + ++L+D+
Sbjct 148 IFGTIINSLGYSNKKKLQQMTRELQEWKEKEESKMSYLRQQVEQLSDK 195
Lambda K H a alpha
0.323 0.138 0.422 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1351240338396