bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-9_CDS_annotation_glimmer3.pl_2_6
Length=529
Score E
Sequences producing significant alignments: (Bits) Value
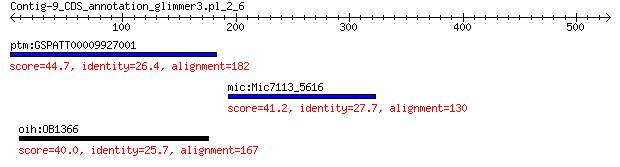
ptm:GSPATT00009927001 hypothetical protein 44.7 0.045
mic:Mic7113_5616 hypothetical protein 41.2 0.71
oih:OB1366 hypothetical protein 40.0 0.91
> ptm:GSPATT00009927001 hypothetical protein
Length=519
Score = 44.7 bits (104), Expect = 0.045, Method: Compositional matrix adjust.
Identities = 48/195 (25%), Positives = 79/195 (41%), Gaps = 26/195 (13%)
Query 1 LNTAAFARM--REYYDFYFVPYDLLWN---KANTSLTQMYDNPQHALDSSPLNVVKLDGS 55
+N A R+ RE + + D+L+ K L Q+ D Q LDS N K+D
Sbjct 183 MNDATLKRVITREEQEILQI-QDVLYQRISKLQQKLIQLNDIEQKQLDSKMNNNNKVDQI 241
Query 56 MPFTDLSSISRYLNSLASNSTAVTNKANYFGYNRALCSAKLMECLGY--GNLYYYAESTS 113
DL + N L NK + A L++CLGY ++YY +
Sbjct 242 QDREDL-----FENQLDEPKQQEQNKGSS-----QQIRAMLIQCLGYQVSSIYYDKKRYQ 291
Query 114 NTFAKRPLHYNLNVSLFNL------LAYQKIYADYYRDSQWERVSPSCFNVDYMPYSTGS 167
TF + N+ + N L Q I+ ++YR+ Q R+ S +DY + +
Sbjct 292 ITFTYK--QANVQYTFQNQKQIDGNLEIQDIFNEFYRELQENRLVLSGLKIDYKQKTEIN 349
Query 168 SGMMLSFNYSDFYEN 182
G ++ + ++N
Sbjct 350 KGEIVQLSLKHQFQN 364
> mic:Mic7113_5616 hypothetical protein
Length=1041
Score = 41.2 bits (95), Expect = 0.71, Method: Compositional matrix adjust.
Identities = 36/135 (27%), Positives = 61/135 (45%), Gaps = 13/135 (10%)
Query 193 WQKDLFHGVVPNQQYGDVASI--SMSVPVVAGSSAALINSRITSSNS---TTTLRFPTDP 247
W + + VP+ +Y D+ + S + + S A I + I N T LR+ P
Sbjct 625 WLRYGWSDTVPSPKYQDIQKLLNSHTAAIYWHVSPAAITTFILRQNKSLKTFVLRYDQPP 684
Query 248 AIPDATPLLTHPSFSILALRQAEFLQKWKEITQSGNKDYKEQVEKHWNVSPGDGFSEMCT 307
+ A +L + +++ A R+ + L+ W + KDYK Q EK W E+ T
Sbjct 685 HLLCAETMLDNSTYAAAA-RKLQDLENWLTHWKQDYKDYKVQGEKRWR-------QEIPT 736
Query 308 YLGGISSSLDINEVV 322
L +S L+I E++
Sbjct 737 RLNELSQILNIAEIL 751
> oih:OB1366 hypothetical protein
Length=298
Score = 40.0 bits (92), Expect = 0.91, Method: Compositional matrix adjust.
Identities = 43/183 (23%), Positives = 80/183 (44%), Gaps = 24/183 (13%)
Query 9 MREYYDFYFVPYDLLWNK-ANTSLTQMY--DNPQHALDSSPLNVVKLDGSMPFTDLSSIS 65
M+ YY F+ + LL+ K ++ ++ +Y +N + LD LN +L G+ F DL ++
Sbjct 1 MKVYYFIIFICFILLFPKVSDAAIIDLYIDENVRGNLD---LNQYQLLGNEGFFDLQKLN 57
Query 66 RYLNSLASNSTAVTNKANYFGYNRALCSAKLMECLGYG-----------NLYYYAESTSN 114
+++ L A Y + + ME +GY YY +
Sbjct 58 TFMDELQKEVNKEPKDAYYDSTGKLI-----MEKVGYTLDREKFIELIYQSYYVENNDKV 112
Query 115 TFAKRPLHYNLNVSLFNLLAYQKI--YADYYRDSQWERVSPSCFNVDYMPYSTGSSGMML 172
T KRP+H +N +L ++ ++I Y +R+S +R + + + + G +
Sbjct 113 TVPKRPVHARVNHALLREISQKQIGFYTTTFRESNTQRANNIKLAAESINNTVLFPGEVF 172
Query 173 SFN 175
SFN
Sbjct 173 SFN 175
Lambda K H a alpha
0.317 0.132 0.400 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1173928257792