bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
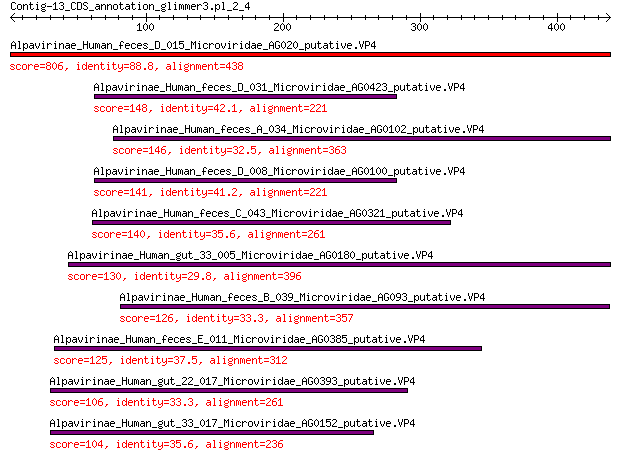
Query= Contig-13_CDS_annotation_glimmer3.pl_2_4
Length=438
Score E
Sequences producing significant alignments: (Bits) Value
Alpavirinae_Human_feces_D_015_Microviridae_AG020_putative.VP4 806 0.0
Alpavirinae_Human_feces_D_031_Microviridae_AG0423_putative.VP4 148 5e-41
Alpavirinae_Human_feces_A_034_Microviridae_AG0102_putative.VP4 146 3e-40
Alpavirinae_Human_feces_D_008_Microviridae_AG0100_putative.VP4 141 1e-38
Alpavirinae_Human_feces_C_043_Microviridae_AG0321_putative.VP4 140 3e-38
Alpavirinae_Human_gut_33_005_Microviridae_AG0180_putative.VP4 130 5e-35
Alpavirinae_Human_feces_B_039_Microviridae_AG093_putative.VP4 126 2e-33
Alpavirinae_Human_feces_E_011_Microviridae_AG0385_putative.VP4 125 4e-33
Alpavirinae_Human_gut_22_017_Microviridae_AG0393_putative.VP4 106 2e-26
Alpavirinae_Human_gut_33_017_Microviridae_AG0152_putative.VP4 104 7e-26
> Alpavirinae_Human_feces_D_015_Microviridae_AG020_putative.VP4
Length=563
Score = 806 bits (2083), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 389/438 (89%), Positives = 415/438 (95%), Gaps = 0/438 (0%)
Query 1 MLEHLRLLEADRYKALALRYPNFISKFRPFILRSIPRVSKLQNFKDEYFEELVWMLPEIA 60
MLEHLRLLEADRYKALALRYPNFISK RP+ILRSIPRVSKLQNFKDEYFEELVWMLPEIA
Sbjct 126 MLEHLRLLEADRYKALALRYPNFISKARPYILRSIPRVSKLQNFKDEYFEELVWMLPEIA 185
Query 61 ESLKKKNNTDASGAFPQFKGLLKYVNIRDYQLFAKRLRKYLSKKIGKYEKIHSYVVSEYS 120
ESLKKKNNTDA+GAFPQFKGLLKYVNIRDYQLFAKRLRKYLSKK+GKYEKIHSYVVSEYS
Sbjct 186 ESLKKKNNTDANGAFPQFKGLLKYVNIRDYQLFAKRLRKYLSKKVGKYEKIHSYVVSEYS 245
Query 121 PKTFRPHFHILFFFDSDEIAKNFRQAVYQSWRLGRVDTQLAREQANSYVANYLNSVVSIP 180
PKTFRPHFHILFFFDSDEIAKNFRQAVYQSWRLGRVDTQLAREQANSYV+NYLNSVVSIP
Sbjct 246 PKTFRPHFHILFFFDSDEIAKNFRQAVYQSWRLGRVDTQLAREQANSYVSNYLNSVVSIP 305
Query 181 FVYKAKKSIRPRSRFSNLFGYEEVKKGIQHASDKRSALFDGVPYISNQKFVRYVPSGSHI 240
FVYKAKKSIRPRSRFSNLFG+EEVK+GI+ A DKR+ALFDG+ YISNQKFVRYVPSGS I
Sbjct 306 FVYKAKKSIRPRSRFSNLFGFEEVKEGIRKAQDKRAALFDGLSYISNQKFVRYVPSGSLI 365
Query 241 DRLFPRFTHYDGSFLRRSSQIYEVVQRVLRLFARNEPFKKATPRNVSEFVCWWCEYNFRR 300
DRLFPRFT+YDGSFLRRSSQIY VVQ++LRLFARNEPF++ATP NVSEF+CWWC YNFR+
Sbjct 366 DRLFPRFTYYDGSFLRRSSQIYGVVQQILRLFARNEPFEEATPGNVSEFICWWCAYNFRQ 425
Query 301 GCQIKDFPDYMQEFLHIVRLDGYSFLNWDVPIGKISRFFYRFNRFEAMKGSLRSKLKAVS 360
GCQIKDFPDYMQEFLHIVRLD SFLNWD+P GKISRF YRFN FE MKGS RSKLKAV
Sbjct 426 GCQIKDFPDYMQEFLHIVRLDRESFLNWDIPKGKISRFLYRFNMFEKMKGSFRSKLKAVE 485
Query 361 LFYDYRDYQSLKNQLSLQELVFAELGYSDELFDSFYVKPHIKVLKNAYIDKWKDVNYKEV 420
LFYDYRDYQSLKNQL +Q+LVF+ELGYSDEL DSFYVKP++KVLKN Y +KW D NY EV
Sbjct 486 LFYDYRDYQSLKNQLYMQQLVFSELGYSDELLDSFYVKPNLKVLKNIYTEKWHDTNYHEV 545
Query 421 HYFRVKHKVLNDENNIFL 438
HYFRVKHKVLND+N++FL
Sbjct 546 HYFRVKHKVLNDQNDVFL 563
> Alpavirinae_Human_feces_D_031_Microviridae_AG0423_putative.VP4
Length=532
Score = 148 bits (374), Expect = 5e-41, Method: Compositional matrix adjust.
Identities = 93/227 (41%), Positives = 126/227 (56%), Gaps = 16/227 (7%)
Query 62 SLKKKNNTDASGAFPQFKGLLKYVNIRDYQLFAKRLRKYLSKKIGKYEKIHSYVVSEYSP 121
S +K N +G +P G + Y+ D L+ KR+RKY+SK +G E IH+Y+V EY P
Sbjct 137 SYAQKANLSFNGKYPALSGRIPYLLHGDVSLYMKRVRKYISK-LGINETIHTYIVGEYGP 195
Query 122 KTFRPHFHILFFFDSDEIAKNFRQAVYQSWRLGRVDTQLAREQANSYVANYLNSVVSIPF 181
+FRPHFH+L FFDSDE+A+N + WR GRVD +R A YV++YLNS SIP
Sbjct 196 SSFRPHFHLLLFFDSDELAQNIIRIASSCWRFGRVDCSASRGDAEDYVSSYLNSFSSIPL 255
Query 182 VYKAKKSIRPRSRFSNLFGYEEVKKGIQHASDKRSALFDGVPYISNQKFVRYVPSGSHID 241
+ ++IRP +RFSN FGY + I+ A +S FD I N K +P
Sbjct 256 HIQEIRAIRPFARFSNKFGYSFFESSIKKA---QSGNFDE---ILNGK---SLPYNGFDT 306
Query 242 RLFPRFTHYDGSF----LRRSSQIYEVVQ--RVLRLFARNEPFKKAT 282
+FP D F LRR S I+E+ + R R F + F+KAT
Sbjct 307 TIFPWRAIIDTCFYRPALRRHSDIHELTEILRYARNFKQRPAFQKAT 353
> Alpavirinae_Human_feces_A_034_Microviridae_AG0102_putative.VP4
Length=539
Score = 146 bits (368), Expect = 3e-40, Method: Compositional matrix adjust.
Identities = 118/399 (30%), Positives = 182/399 (46%), Gaps = 52/399 (13%)
Query 76 PQFKGLLKYVNIRDYQLFAKRLRKYLSKKIGKYEKIHSYVVSEYSPKTFRPHFHILFFFD 135
P +GL+ Y+N D QLF KRL + + + EKI+ YVV EY P TFRPHFHIL F D
Sbjct 149 PYMEGLVGYLNYHDIQLFFKRLNQNIRRITN--EKIYYYVVGEYGPTTFRPHFHILLFHD 206
Query 136 SDEIAKNFRQAVYQSWRLGRVDTQLAREQANSYVANYLNSVVSIPFVYKAKKSIRPRSRF 195
S ++ ++ RQ V +SWR G DTQ A+ YVA Y+NS +P +K + I+P RF
Sbjct 207 SRKLRESIRQFVSKSWRFGDSDTQPVWSSASCYVAGYVNSTACLPDFFKNSRHIKPFGRF 266
Query 196 SNLF---GYEEVKKGIQHASDKRSALFDGVPYISNQKFVRYVPSGSHIDRLFPRFTHYDG 252
S F + EV K + + S +DG N K P SHI+RL+PR
Sbjct 267 SVNFAESAFNEVFKP-EENEEIFSLFYDGRVLNLNGKPTIVRPKRSHINRLYPRLDKSKH 325
Query 253 SFLRRSSQIYEVVQRVLRLFARNEPFKKATPRNVSEFVCWWCEYNFRRGCQIKDFPDYMQ 312
+ + +I VV R+ ++ A+ + + +S+ V + + C + D ++
Sbjct 326 ATVVDDIRIASVVSRLPQVVAKFGFIDEVSDFELSKRVYYLIRRHLEVDCSLDYASDELR 385
Query 313 EFLHIVRLDGYSFLNWDVPIGKISRFFYRFNRF-----------EAMKGSLRSKLKAVSL 361
+ RL Y + + I R ++ A G L ++A+
Sbjct 386 VIYNACRLSLYINFSDESGCAAIYRLLLQYRNLVKNWITAPVGSVAFTGQLHRAVRAIHS 445
Query 362 FYDYRDYQSLKNQL----------------------SLQELVFAELGYSDELFDSFYVKP 399
FYDY +SL +QL L ++ +S+ +FDS
Sbjct 446 FYDYCSRRSLHDQLVKVEKWSNDSYVRANLSIYYFYPLTDVDIMMKSFSEIVFDS----- 500
Query 400 HIKVLKNAYIDKWKDVNYKEVHYFRVKHKVLNDENNIFL 438
+VL+ +Y D Y + R+KHK LND+N++ +
Sbjct 501 --QVLRASYAD------YAADNRERIKHKALNDKNSLLI 531
> Alpavirinae_Human_feces_D_008_Microviridae_AG0100_putative.VP4
Length=532
Score = 141 bits (355), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 91/227 (40%), Positives = 125/227 (55%), Gaps = 16/227 (7%)
Query 62 SLKKKNNTDASGAFPQFKGLLKYVNIRDYQLFAKRLRKYLSKKIGKYEKIHSYVVSEYSP 121
S +K N G +P + Y+ D L+ KR+RKY+SK +G E IH+YVV EY P
Sbjct 137 SYAQKANLSFDGKYPALSDRIPYLLHDDVSLYMKRVRKYISK-LGINETIHTYVVGEYGP 195
Query 122 KTFRPHFHILFFFDSDEIAKNFRQAVYQSWRLGRVDTQLAREQANSYVANYLNSVVSIPF 181
TFRPHFH+L FF+SDE+A++ + WR GRVD +R A YV++YLNS SIP
Sbjct 196 VTFRPHFHLLLFFNSDELAQSIVRIARSCWRFGRVDCSASRGDAEDYVSSYLNSFSSIPL 255
Query 182 VYKAKKSIRPRSRFSNLFGYEEVKKGIQHASDKRSALFDGVPYISNQKFVRYVPSGSHID 241
+ ++IRP +RFSN FG+ + I+ A +S FD I N K +P
Sbjct 256 HIQEIRAIRPFARFSNKFGFSFFESSIKKA---QSGDFDE---ILNGK---SLPYNGFNT 306
Query 242 RLFPRFTHYDGSF----LRRSSQIYEVVQ--RVLRLFARNEPFKKAT 282
+FP D F LRR S I+E+ + R +R F + F+KAT
Sbjct 307 TIFPWRAIIDTCFYRPALRRRSDIHELTEILRYVRNFKQRPAFQKAT 353
> Alpavirinae_Human_feces_C_043_Microviridae_AG0321_putative.VP4
Length=546
Score = 140 bits (352), Expect = 3e-38, Method: Compositional matrix adjust.
Identities = 93/275 (34%), Positives = 147/275 (53%), Gaps = 19/275 (7%)
Query 61 ESLKKKNNTDASGAFPQFKGLLKYVNIRDYQLFAKRLRKYLSKKIGKYEKIHSYVVSEYS 120
E+ KK+ N D G +P K + Y++ +D QLF KR+RK + EKIH+Y+V EYS
Sbjct 131 ENFKKQANLDVKGCYPHLKDMYGYLSRKDCQLFMKRVRKQIRNYTD--EKIHTYIVGEYS 188
Query 121 PKTFRPHFHILFFFDSDEIAKNFRQAVYQSWRLGRVDTQLAREQANSYVANYLNSVVSIP 180
PK FRPHFHILFFF+S+E++++F V W+ G VD +R A SYVA Y+NS +P
Sbjct 189 PKHFRPHFHILFFFNSNELSQSFGSIVRSCWKFGLVDWSQSRGDAESYVAGYVNSFARLP 248
Query 181 FVYKAKKSIRPRSRFSNLFG---YEEVKKGIQHA-SDKRSALF----DGVPYISNQKFVR 232
+ + I P RFSN F +++ K+ ++ + +++ +F +GV + N K +
Sbjct 249 YHLGSSPKIAPFGRFSNHFSESCFDDAKQTLRDSFFGQKTPVFAPFLNGVTNLVNGKLLL 308
Query 233 YVPSGSHIDRLFPRFTHYDG-----SFLRRSSQIYEVVQRVLRLFARNEPFKKATPRNVS 287
PS S +D F R DG L + VV++ R FAR+ T N +
Sbjct 309 TRPSRSCVDSCFFRKAR-DGRLSSHELLHLIRAVCHVVEQGKRFFARDG--LNYTFLNHA 365
Query 288 EFVCWWCE-YNFRRGCQIKDFPDYMQEFLHIVRLD 321
F+ C+ + +++ ++ P + L+ R+D
Sbjct 366 RFIVKACKSFRYQKREKLLSLPSRLVTLLYYSRVD 400
> Alpavirinae_Human_gut_33_005_Microviridae_AG0180_putative.VP4
Length=536
Score = 130 bits (327), Expect = 5e-35, Method: Compositional matrix adjust.
Identities = 118/429 (28%), Positives = 193/429 (45%), Gaps = 38/429 (9%)
Query 43 NFKDEYFEELVWMLPEIAESLKKKNNTDASG--------AFPQFKGLLKYVNIRDYQLFA 94
+F DE F+ + E +L K + D + +P LL Y+N RD QLF
Sbjct 107 SFCDEEFDYPTNLRDEAVTALLDKTHLDRTVYPDGRSVVKYPNMGDLLPYLNYRDVQLFH 166
Query 95 KRLRKYLSKKIGKY--EKIHSYVVSEYSPKTFRPHFHILFFFDSDEIAKNFRQAVYQSWR 152
KR+ +++I KY EKI+SY V EY PKTFRPHFH+LFFFDS+ +A+ F Q V ++WR
Sbjct 167 KRI----NQQIKKYTDEKIYSYTVGEYGPKTFRPHFHLLFFFDSERLAQVFGQLVDKAWR 222
Query 153 LGRVDTQLAREQANSYVANYLNSVVSIPFVYKAKKSIRPRSRFSNLFGYEEVKKGIQHAS 212
G DTQ A+SYVA YLNS +P Y+ + I P RFS F + +
Sbjct 223 FGNSDTQRVWSSASSYVAGYLNSSHCLPEFYRCNRKIAPFGRFSQYFAERPFIEAFKPEE 282
Query 213 DKR--SALFDGVPYISNQKFVRYVPSGSHIDRLFPRFTHYDGSFLRRSSQIYEVVQRVLR 270
+++ +G+ K P S I+RL+P S + + + V ++ +
Sbjct 283 NEKVFDKFVNGIYLSLGGKPTLCRPKRSLINRLYPVLDRSAVSDVDSNVRTALFVSKIPQ 342
Query 271 LFARNEPFKKATPRNVSEFVCWWCEYNFRRGCQIKDFPDYMQEFLHIVRLDGYSFLNWDV 330
+ A+ + T ++ + + + + + P+ ++ + RL Y +
Sbjct 343 VLAKFGFLDEVTLFEQAKRTFFVIKKYLQVDHSLDNAPETLRFIYNACRLSLYLNFSELE 402
Query 331 PIGKISRFFYRFNRFE-----------AMKGSLRSKLKAVSLFYDYRDYQSLKNQLSLQE 379
+ R F + A G LR + + FY Y D + L +QL ++
Sbjct 403 GCSAVYRLFLAYRNLCDHWITVPSNHIAFYGQLRKAFRTIYAFYSYMDSKYLHDQL-VKV 461
Query 380 LVFAELGYSDELFDSFYVKPHI--KVLKNAY-----IDKWKDVNYKEV---HYFRVKHKV 429
+++ Y D Y P ++++++Y + + Y EV + R+KHK
Sbjct 462 QSWSQNNYLSTQVDFRYFYPLTDGELMRSSYREVLALSSFCRAAYAEVAADNRQRIKHKY 521
Query 430 LNDENNIFL 438
LND N +
Sbjct 522 LNDLNCALI 530
> Alpavirinae_Human_feces_B_039_Microviridae_AG093_putative.VP4
Length=559
Score = 126 bits (316), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 119/388 (31%), Positives = 185/388 (48%), Gaps = 42/388 (11%)
Query 81 LLKYVNIRDYQLFAKRLRKYLSKKIGKYEKIHSYVVSEYSPKTFRPHFHILFFFDSDEIA 140
L+ ++N D Q + KRLRKYL +++GKYE H Y V EY P FRPH+H+L F +SD+++
Sbjct 179 LIPFLNYVDVQNYIKRLRKYLFQQLGKYETFHFYAVGEYGPVHFRPHYHLLLFTNSDKVS 238
Query 141 KNFRQAVYQSWRLGRVDTQLAREQANSYVANYLNSVVSIPFVYKAKKSIRPRSRFSNLF- 199
+ R +SW+LGR D Q + A SYVA+Y+NS+ S P +Y++ ++ RP+SR S F
Sbjct 239 EVLRYCHDKSWKLGRSDFQRSAGGAGSYVASYVNSLCSAPLLYRSCRAFRPKSRASVGFF 298
Query 200 --GYEEVKKGIQHA--SDKRSALFDGVPYISNQKFVRYVPSGSHIDRLFPRFT---HYDG 252
G + V+ +A K ++ +G Y N VR P S+I L PRF+ + DG
Sbjct 299 EKGCDFVEDDDPYAQIEQKIDSVVNGRVYNFNGVSVRSTPPLSYIRTLLPRFSSARNDDG 358
Query 253 SFLRRSSQIYEVVQRVLRLFARNEPFKKATPRNVSEFVCWWCEYNFRRGCQIKDFPDYMQ 312
+ R I V + AR F A + S Y + + I D +
Sbjct 359 IAIAR---ILYAVHSTPKRIAR---FGFADYKQDSILSLVRTYYQYLKVNPI--LTDDDK 410
Query 313 EFLHIVR-----LDGYSFLNWDVPIGKISRFF---YRFNR----------FEAMKGSLRS 354
LH R ++ S ++ + I K+ R F Y+F R A G +
Sbjct 411 LILHASRCLTRFVNCSSDVDIESYINKLYRLFLYVYKFFRNWHLPFFGSDISAFSGRIMF 470
Query 355 KLKAVSLFYDYRDYQSLKNQLSLQELVFAELGYSDELFDSFYVKPHIKVLKNAYIDK--- 411
+K + +DY+SL++ +++ A SD +F VL + +
Sbjct 471 IIKTGIEYEKKKDYESLRDVFNIRS---ANPNISDCMFALPANGQERDVLSDVSCETVQL 527
Query 412 WKDVNYKEVHYFR--VKHKVLNDENNIF 437
+ + + + R +KHK LND NNIF
Sbjct 528 LEQLRLRSATFCRDMIKHKRLNDANNIF 555
> Alpavirinae_Human_feces_E_011_Microviridae_AG0385_putative.VP4
Length=611
Score = 125 bits (315), Expect = 4e-33, Method: Compositional matrix adjust.
Identities = 117/352 (33%), Positives = 168/352 (48%), Gaps = 45/352 (13%)
Query 33 RSIPR-VS-------KLQNFKDEYFEELVWM-LPEIAESLKKKNNTDASG-------AFP 76
+SIPR VS + ++F DE + + M L E+ + L K N G +
Sbjct 136 QSIPRDVSVKSKGSFRFRSFDDEPLKFCIPMKLTELQDILIKANGRYDYGKKKVVYPSLA 195
Query 77 QFKGLLKYVNIRDYQLFAKRLRKYLSKKIGKYEKIHSYVVSEYSPKTFRPHFHILFFFDS 136
K + + RD +LF KRLR+ L KI YVVSEY P+T+RPH+H L FF+S
Sbjct 196 DCKLQIPVLQSRDIELFFKRLRRNLDSHGFTSSKICYYVVSEYGPQTYRPHWHCLLFFNS 255
Query 137 DEIAKNFRQAVYQSWRLGRVDTQLAREQANSYVANYLNSVVSIPFVYKAKKSIRPRSRFS 196
+EI + R+ + ++W GR+D L+R A SYVA Y+NS +PF+Y +K IRPRS S
Sbjct 256 EEITQTLREDISKAWSYGRIDYSLSRGAAASYVAAYVNSAACLPFLYVGQKEIRPRSFHS 315
Query 197 NLFGYEEV---KKGIQHASDKRSALFDGVPYISNQKFVRYVPSGSHIDRLFPRFTHYDGS 253
FG +V + S FDGV SN K V P S +FPRF++
Sbjct 316 KGFGSNKVFPKSSDVSEISKISDLFFDGVNVDSNGKVVNIRPVRSCELAVFPRFSN---D 372
Query 254 FLRRSSQIYEVVQRVL----RLFAR-----NEPFKKATPRNVSEFVCWWCEY-------- 296
F S ++ Q V+ RL +R + P + +S+ V + EY
Sbjct 373 FFSDSDTCCKLFQSVIETPERLVSRGYLGIDTPNFGSDDFRLSDLVRAYSEYYERNFTDF 432
Query 297 ----NFRRGCQIKDFPDYMQEFLHIVRLDGYSFLNWDVPIGKISRFFYRFNR 344
F RG + +D+ D + F DGY F N D+ G++ R F + R
Sbjct 433 SFSLRFLRGYRTRDYADELI-FREARLFDGYVF-NKDMIFGRLYRLFAKVLR 482
> Alpavirinae_Human_gut_22_017_Microviridae_AG0393_putative.VP4
Length=562
Score = 106 bits (264), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 87/286 (30%), Positives = 138/286 (48%), Gaps = 43/286 (15%)
Query 30 FILRSIPRVSKLQ--------NFKDEYFEELVWMLPEIAESLKKKNNTDASGAFPQFKGL 81
F RS+PR S ++ FKD + M P+ S+ K N + P
Sbjct 110 FGFRSVPRSSSVKLKSSTVERTFKDPDVKFSYPMKPKELLSILGKINHNVPNRIP----- 164
Query 82 LKYVNIRDYQLFAKRLRKYLSKKIGKYEKIHSYVVSEYSPKTFRPHFHILFFFDSDEIAK 141
Y+ RD LF KRLR Y EK+ Y VSEY P +FRPH+H+L F +S+ ++
Sbjct 165 --YICNRDLDLFLKRLRSYYLD-----EKLRYYAVSEYGPTSFRPHWHLLLFSNSERFSR 217
Query 142 NFRQAVYQSWRLGRVDTQLAREQANSYVANYLNSVVSIP-FVYKAKKSIRPRS------R 194
+ V ++W GR D L+R A YVA+Y+NS V++P F + K +RP+S
Sbjct 218 TVCENVSKAWSYGRCDASLSRGFAAPYVASYVNSFVALPDFYTQMPKVVRPKSFHSIGFT 277
Query 195 FSNLFGYEEVKKGIQHASDKRSALFDGVPYISNQKFVRYVPSGSHIDRLFPRFTHYDGSF 254
SNLF + + +DK +GV + F P+ ++ RLFPRF+ +
Sbjct 278 ESNLFPRKVRVAEVDEITDK---CLNGVRVERDGYFRTIKPTWPYLLRLFPRFS---DAI 331
Query 255 LRRSSQIYEVV-------QRVLRLFARN---EPFKKATPRNVSEFV 290
+ S IY+++ +RV+R + +PF +++ +++ F
Sbjct 332 RKSPSSIYQLLFAAFTAPERVIRSGCADIGCDPFGESSKQSILSFC 377
> Alpavirinae_Human_gut_33_017_Microviridae_AG0152_putative.VP4
Length=565
Score = 104 bits (259), Expect = 7e-26, Method: Compositional matrix adjust.
Identities = 84/251 (33%), Positives = 124/251 (49%), Gaps = 33/251 (13%)
Query 30 FILRSIPRVS--KLQN------FKDEYFEELVWMLPEIAESLKKKNNTDASGAFPQFKGL 81
F S+PR + KL+N FKD M P+ S+ K N + P
Sbjct 110 FGFCSVPRSASVKLKNSTVERTFKDPEVRFSYPMKPKELLSILDKINHNVPNRIP----- 164
Query 82 LKYVNIRDYQLFAKRLRKYLSKKIGKYEKIHSYVVSEYSPKTFRPHFHILFFFDSDEIAK 141
Y+ RD LF KRLR Y YEK+ Y VSEY P ++RPH+H+L F +S++ +K
Sbjct 165 --YICNRDLDLFLKRLRSYY-----PYEKLRYYAVSEYGPTSYRPHWHLLLFSNSEQFSK 217
Query 142 NFRQAVYQSWRLGRVDTQLAREQANSYVANYLNSVVSIP-FVYKAKKSIRPRS------R 194
+ V ++W GR D L+R A YVA+Y+NS V++P F + + +RP+S
Sbjct 218 TILENVSKAWSYGRCDASLSRGFAAPYVASYVNSFVALPSFYTEMPRFLRPKSFHSIGFT 277
Query 195 FSNLFGYEEVKKGIQHASDKRSALFDGVPYISNQKFVRYVPSGSHIDRLFPRFTHYDGSF 254
SNLF + I +DK +GV + F PS ++ RLFPRF+ +
Sbjct 278 ESNLFPRKVRISEIDEVTDK---CLNGVRVERDGYFRILKPSWPYLLRLFPRFS---DAI 331
Query 255 LRRSSQIYEVV 265
+ S IY+++
Sbjct 332 RKSPSSIYQLL 342
Lambda K H a alpha
0.325 0.140 0.426 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 39128072