bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-1_CDS_annotation_glimmer3.pl_2_1
Length=289
Score E
Sequences producing significant alignments: (Bits) Value
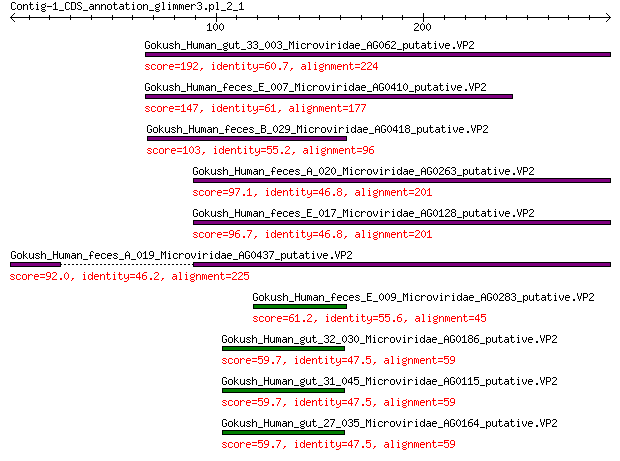
Gokush_Human_gut_33_003_Microviridae_AG062_putative.VP2 192 2e-60
Gokush_Human_feces_E_007_Microviridae_AG0410_putative.VP2 147 1e-43
Gokush_Human_feces_B_029_Microviridae_AG0418_putative.VP2 103 2e-27
Gokush_Human_feces_A_020_Microviridae_AG0263_putative.VP2 97.1 4e-25
Gokush_Human_feces_E_017_Microviridae_AG0128_putative.VP2 96.7 5e-25
Gokush_Human_feces_A_019_Microviridae_AG0437_putative.VP2 92.0 2e-23
Gokush_Human_feces_E_009_Microviridae_AG0283_putative.VP2 61.2 1e-12
Gokush_Human_gut_32_030_Microviridae_AG0186_putative.VP2 59.7 4e-12
Gokush_Human_gut_31_045_Microviridae_AG0115_putative.VP2 59.7 4e-12
Gokush_Human_gut_27_035_Microviridae_AG0164_putative.VP2 59.7 4e-12
> Gokush_Human_gut_33_003_Microviridae_AG062_putative.VP2
Length=295
Score = 192 bits (487), Expect = 2e-60, Method: Compositional matrix adjust.
Identities = 136/227 (60%), Positives = 161/227 (71%), Gaps = 4/227 (2%)
Query 66 TNVQQVNDWLKQAYAYQGQEAAMQGKYNSQSMLKQMGYNTLQAIMQGVYNHIENSVAMNY 125
TN +Q+ +L + YA+QG + A Q K N Q+ML QMGYNTL AI QG+YNHIE + AMNY
Sbjct 70 TNDEQIMKYLDRFYAWQGGQNAFQSKTNRQNMLMQMGYNTLGAIQQGIYNHIEQNAAMNY 129
Query 126 NSTEALANREWQEHMSSTAYQRAVEDMKKAGLNPILAFsnggastpggsagtisgasmgl 185
NS EALANR +QE MSST+YQRAVEDM+KAGLNPILAF+NGGASTPGGS TI+GASMG+
Sbjct 130 NSAEALANRNFQERMSSTSYQRAVEDMRKAGLNPILAFANGGASTPGGSGATITGASMGM 189
Query 186 asssalgvsrsggFVPNAYsssswsksDWYNAAQSWQQMLSTTQMTPYGLMKTLTKV--- 242
SSSALGVS G VP + S S S + WY A++ LST+ TP L+ L K
Sbjct 190 PSSSALGVSTMNGNVPTSNYSRSESNAQWYQLAEAVGSQLSTSHSTPKALVDDLLKTYKA 249
Query 243 GSDTSEAIKDATAKTEKGKGAEQDRSIKPQDKTGSYGEKRKPGDYLK 289
T E + A + K +QDR+IKPQDKTG YGEKRKPGDYLK
Sbjct 250 MEKTEETVPGAAGGGGRSK-TKQDRAIKPQDKTGKYGEKRKPGDYLK 295
> Gokush_Human_feces_E_007_Microviridae_AG0410_putative.VP2
Length=270
Score = 147 bits (371), Expect = 1e-43, Method: Compositional matrix adjust.
Identities = 108/177 (61%), Positives = 132/177 (75%), Gaps = 0/177 (0%)
Query 66 TNVQQVNDWLKQAYAYQGQEAAMQGKYNSQSMLKQMGYNTLQAIMQGVYNHIENSVAMNY 125
TN +Q+ D+L + Y +QG + A Q K N Q+ML QMGYNTL AI QG+YNHIEN+ AM Y
Sbjct 67 TNDKQIMDYLNRYYQWQGGQNAFQSKTNRQNMLMQMGYNTLSAIQQGIYNHIENNAAMQY 126
Query 126 NSTEALANREWQEHMSSTAYQRAVEDMKKAGLNPILAFsnggastpggsagtisgasmgl 185
NS EALANR++QE MSSTAYQRAVEDM+KAGLNPILA++ GGASTPGGS TI+GASMG+
Sbjct 127 NSAEALANRQFQERMSSTAYQRAVEDMRKAGLNPILAYAQGGASTPGGSGATITGASMGM 186
Query 186 asssalgvsrsggFVPNAYsssswsksDWYNAAQSWQQMLSTTQMTPYGLMKTLTKV 242
+SSALGVS G VPN+Y + S SKS WY A++ +ST +P L + L K
Sbjct 187 PTSSALGVSTLSGNVPNSYFNRSESKSQWYQLAEAVGSQMSTGYSSPVQLTEDLLKT 243
> Gokush_Human_feces_B_029_Microviridae_AG0418_putative.VP2
Length=272
Score = 103 bits (257), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 53/96 (55%), Positives = 63/96 (66%), Gaps = 0/96 (0%)
Query 67 NVQQVNDWLKQAYAYQGQEAAMQGKYNSQSMLKQMGYNTLQAIMQGVYNHIENSVAMNYN 126
NV Q D K Q MQ N+++ L +G NTL AI QGVYN I+ AM+YN
Sbjct 67 NVDQTKDLAKYFLGQSQQAQGMQSLQNNKNSLMALGLNTLGAIQQGVYNRIQQDAAMSYN 126
Query 127 STEALANREWQEHMSSTAYQRAVEDMKKAGLNPILA 162
S EA ANR WQE MS+T+YQRA EDM+KAG+NPILA
Sbjct 127 SAEAAANRAWQERMSNTSYQRATEDMRKAGINPILA 162
> Gokush_Human_feces_A_020_Microviridae_AG0263_putative.VP2
Length=300
Score = 97.1 bits (240), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 94/206 (46%), Positives = 118/206 (57%), Gaps = 10/206 (5%)
Query 89 QGKYNSQSMLKQMGYNTLQAIMQGVYNHIENSVAMNYNSTEALANREWQEHMSSTAYQRA 148
QG YN Q N++ QG N AM YNS EA NREWQEHMSSTAYQRA
Sbjct 100 QGIYNMAGSGFQGLLNSMMMNKQGSMNAALMREAMAYNSAEAALNREWQEHMSSTAYQRA 159
Query 149 VEDMKKAGLNPILAFsnggastpggsagtisgasmg--lasssalgvsrsggFVPNAYss 206
V DM+ AG+NPILA NGGA+ GGSAG++ GAS+G S++++ S F N+Y
Sbjct 160 VADMRAAGINPILAALNGGAAMGGGSAGSVGGASVGLGSTSAASISALGSPAFSSNSY-- 217
Query 207 sswsksDWYNAAQ---SWQQMLSTTQMTPYGLMKTLTKVGSDTSEAIKDATAKTEKGKGA 263
S SD N AQ S+ Q + + + L K + V +E DA K +K
Sbjct 218 --GSFSDSINLAQGVSSYFQQGANSGKSWQELKKNVEVVTDKVAEPSYDA-GKAQKRVAG 274
Query 264 EQDRSIKPQDKTGSYGEKRKPGDYLK 289
DR+IK DKTGSYG++RKPGDYL+
Sbjct 275 STDRAIKATDKTGSYGQQRKPGDYLR 300
> Gokush_Human_feces_E_017_Microviridae_AG0128_putative.VP2
Length=300
Score = 96.7 bits (239), Expect = 5e-25, Method: Compositional matrix adjust.
Identities = 94/206 (46%), Positives = 118/206 (57%), Gaps = 10/206 (5%)
Query 89 QGKYNSQSMLKQMGYNTLQAIMQGVYNHIENSVAMNYNSTEALANREWQEHMSSTAYQRA 148
QG YN Q N++ QG N AM YNS EA NREWQEHMSSTAYQRA
Sbjct 100 QGIYNMAGSGFQGLLNSMMMNKQGRMNADLMREAMAYNSAEAALNREWQEHMSSTAYQRA 159
Query 149 VEDMKKAGLNPILAFsnggastpggsagtisgasmg--lasssalgvsrsggFVPNAYss 206
V DM+ AG+NPILA NGGA+ GGSAG++ GAS+G S++++ S F N+Y
Sbjct 160 VADMRAAGINPILAALNGGAAMGGGSAGSVGGASVGLGSTSAASISALGSPAFSSNSY-- 217
Query 207 sswsksDWYNAAQ---SWQQMLSTTQMTPYGLMKTLTKVGSDTSEAIKDATAKTEKGKGA 263
S SD N AQ S+ Q + + + L K + V +E DA K +K
Sbjct 218 --GSFSDSINLAQGVSSYFQQGANSGKSWQELKKNVEVVTDKVAEPSYDA-GKAQKRVAG 274
Query 264 EQDRSIKPQDKTGSYGEKRKPGDYLK 289
DR+IK DKTGSYG++RKPGDYL+
Sbjct 275 STDRAIKATDKTGSYGQQRKPGDYLR 300
> Gokush_Human_feces_A_019_Microviridae_AG0437_putative.VP2
Length=300
Score = 92.0 bits (227), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 95/213 (45%), Positives = 122/213 (57%), Gaps = 24/213 (11%)
Query 89 QGKYNSQSMLKQMGYNTLQAIMQGVYNHIENSVAMNYNSTEALANREWQEHMSSTAYQRA 148
QG YN Q N++ QG N AM YNS EA NREWQE+MSSTAYQRA
Sbjct 100 QGIYNMAGSGFQGLLNSMMMNKQGRMNAELMREAMAYNSAEAAHNREWQEYMSSTAYQRA 159
Query 149 VEDMKKAGLNPILAFsnggastpggsagtisgasmglasssalgvsrsg--gFVPNAYss 206
V DM+ AG+NPILA NGGA+ GGSAG++ GAS+GL S+SA +S G F N+Y S
Sbjct 160 VADMRAAGINPILAALNGGAAMGGGSAGSVGGASVGLGSTSAASISALGGPAFSSNSYGS 219
Query 207 ssws------ksDWY----NAAQSWQQMLSTTQMTPYGLMKTLTKVGSDTSEAIKDATAK 256
S S S ++ N+ +SWQQ+ + ++ + + K D +A K
Sbjct 220 FSDSINLAQGVSSYFQQGANSGRSWQQLKNNVEV----VTDKVAKPSYDAGKAQKRVAGS 275
Query 257 TEKGKGAEQDRSIKPQDKTGSYGEKRKPGDYLK 289
T DR+IK DKTGSYG+KR+PGDYL+
Sbjct 276 T--------DRAIKATDKTGSYGQKRQPGDYLR 300
Score = 21.9 bits (45), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 9/24 (38%), Positives = 15/24 (63%), Gaps = 0/24 (0%)
Query 1 MGALSGFLTALNVAGNVANTVGTI 24
+G L + A+ GN+A TVG++
Sbjct 3 LGWLPTAIRAIGTIGNIAGTVGSL 26
> Gokush_Human_feces_E_009_Microviridae_AG0283_putative.VP2
Length=294
Score = 61.2 bits (147), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 25/45 (56%), Positives = 36/45 (80%), Gaps = 0/45 (0%)
Query 118 ENSVAMNYNSTEALANREWQEHMSSTAYQRAVEDMKKAGLNPILA 162
+N AM +N+ EA NR+WQE+MS+TA+QR + D+K AGLNP+L+
Sbjct 64 QNRKAMEFNAAEAAKNRDWQEYMSNTAHQREIADLKAAGLNPVLS 108
> Gokush_Human_gut_32_030_Microviridae_AG0186_putative.VP2
Length=289
Score = 59.7 bits (143), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 28/59 (47%), Positives = 38/59 (64%), Gaps = 0/59 (0%)
Query 103 YNTLQAIMQGVYNHIENSVAMNYNSTEALANREWQEHMSSTAYQRAVEDMKKAGLNPIL 161
+N QA +Q + + M +NS EA NR+WQE MS+TA+QR V D+ AGLNP+L
Sbjct 41 FNAEQAKLQRDWTESMTARQMEFNSAEAAKNRQWQEMMSNTAHQREVRDLMAAGLNPVL 99
> Gokush_Human_gut_31_045_Microviridae_AG0115_putative.VP2
Length=289
Score = 59.7 bits (143), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 28/59 (47%), Positives = 38/59 (64%), Gaps = 0/59 (0%)
Query 103 YNTLQAIMQGVYNHIENSVAMNYNSTEALANREWQEHMSSTAYQRAVEDMKKAGLNPIL 161
+N QA +Q + + M +NS EA NR+WQE MS+TA+QR V D+ AGLNP+L
Sbjct 41 FNAEQAKLQRDWTESMTARQMEFNSAEAAKNRQWQEMMSNTAHQREVRDLMAAGLNPVL 99
> Gokush_Human_gut_27_035_Microviridae_AG0164_putative.VP2
Length=289
Score = 59.7 bits (143), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 28/59 (47%), Positives = 38/59 (64%), Gaps = 0/59 (0%)
Query 103 YNTLQAIMQGVYNHIENSVAMNYNSTEALANREWQEHMSSTAYQRAVEDMKKAGLNPIL 161
+N QA +Q + + M +NS EA NR+WQE MS+TA+QR V D+ AGLNP+L
Sbjct 41 FNAEQAKLQRDWTESMTARQMEFNSAEAAKNRQWQEMMSNTAHQREVRDLMAAGLNPVL 99
Lambda K H a alpha
0.310 0.123 0.349 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 23943361