bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
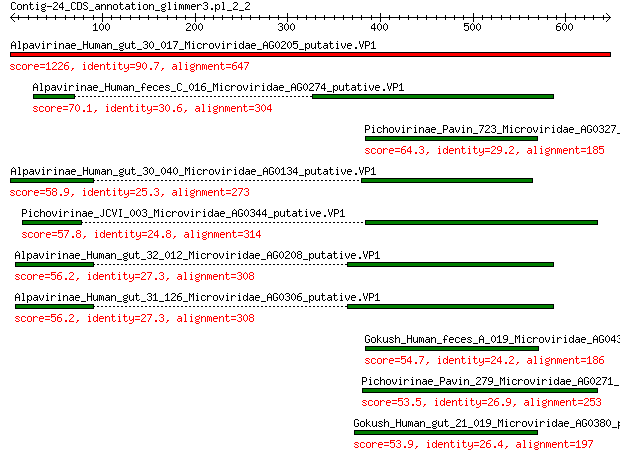
Query= Contig-24_CDS_annotation_glimmer3.pl_2_2
Length=647
Score E
Sequences producing significant alignments: (Bits) Value
Alpavirinae_Human_gut_30_017_Microviridae_AG0205_putative.VP1 1226 0.0
Alpavirinae_Human_feces_C_016_Microviridae_AG0274_putative.VP1 70.1 2e-14
Pichovirinae_Pavin_723_Microviridae_AG0327_putative.VP1 64.3 1e-12
Alpavirinae_Human_gut_30_040_Microviridae_AG0134_putative.VP1 58.9 7e-11
Pichovirinae_JCVI_003_Microviridae_AG0344_putative.VP1 57.8 1e-10
Alpavirinae_Human_gut_32_012_Microviridae_AG0208_putative.VP1 56.2 4e-10
Alpavirinae_Human_gut_31_126_Microviridae_AG0306_putative.VP1 56.2 4e-10
Gokush_Human_feces_A_019_Microviridae_AG0438_putative.VP1 54.7 1e-09
Pichovirinae_Pavin_279_Microviridae_AG0271_putative.VP1 53.5 3e-09
Gokush_Human_gut_21_019_Microviridae_AG0380_putative.VP1 53.9 3e-09
> Alpavirinae_Human_gut_30_017_Microviridae_AG0205_putative.VP1
Length=647
Score = 1226 bits (3171), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 587/647 (91%), Positives = 617/647 (95%), Gaps = 0/647 (0%)
Query 1 MGKQPFISHSVNGYSRYDMPESKAFSITPGIIYPVRIQFVNARDRVTLHQGIDIRSNPLG 60
MGKQPFISHSVNGYSRYDMPE+KAFSITPGIIYPVRIQFVNARDRVTLHQG+D+RSNPLG
Sbjct 1 MGKQPFISHSVNGYSRYDMPENKAFSITPGIIYPVRIQFVNARDRVTLHQGVDVRSNPLG 60
Query 61 VPSFNPYVLRLHRFWVPLQLYHPELRVNSSKFDMNNLTYNFIPGVVDCAGAGSYTSCMYP 120
VPSFNPYVLRLHRFWVPLQLYHPE+RVNSSKFDMNNLTYNFIPGVVD G G+YTS MYP
Sbjct 61 VPSFNPYVLRLHRFWVPLQLYHPEMRVNSSKFDMNNLTYNFIPGVVDNKGGGNYTSYMYP 120
Query 121 RSGTAAFFSQVMPFNHRAALPNSLMSWLRIANSPIFCYSGKPLPVSTCLLKTAPKFLSVN 180
RSGTAAFFSQVMPFN RAALPNSLMSWLRIANSPIF YSG PLP L KT+PKFLSVN
Sbjct 121 RSGTAAFFSQVMPFNCRAALPNSLMSWLRIANSPIFNYSGNPLPSGGVLFKTSPKFLSVN 180
Query 181 ADTYLGYWDIVRNYYSYSSWGVFSFAHPGTYRPVFYTTPTSSVTKVEYRSQASYFWQRYG 240
ADTYLGYWDIVRNYYSYSSWGVFSFAHPGTYRPVFYT +SSVT VEYRSQASYFWQRYG
Sbjct 181 ADTYLGYWDIVRNYYSYSSWGVFSFAHPGTYRPVFYTGTSSSVTNVEYRSQASYFWQRYG 240
Query 241 NLEFLDHYFETMFYPRDRKTPDDRDELSWNRSDLFVEILRSDLFNEDTIAASPDFTNLVQ 300
NLEFLDHYFETMFYPRDR DRDELSWNRSDLFVEILRSDLFN+DT AASPDFT + Q
Sbjct 241 NLEFLDHYFETMFYPRDRIVAADRDELSWNRSDLFVEILRSDLFNKDTTAASPDFTKMPQ 300
Query 301 LFPQGINCRVPAHPYDVQEPKVDWNNGTGTDVNIPSKVYFSATLNVPFLAAHPMAVCPSS 360
++P+ +N VPA+PYDVQEPKVDWNNGTGTDVN+PSKV+F+ATLNVPFLAAHPMAVCPSS
Sbjct 301 MYPEAMNFNVPAYPYDVQEPKVDWNNGTGTDVNVPSKVFFAATLNVPFLAAHPMAVCPSS 360
Query 361 PDRFSRLMPPGDSNSDVDFTGVKTIPQLAVATRLQEYKDLIGASGSRYSDWLYTFFASKI 420
PDRFSRLMPPGDSNSDVDFTG+KTIPQLAVATRLQEYKDLIGASGSRYSDWLYTFFASKI
Sbjct 361 PDRFSRLMPPGDSNSDVDFTGLKTIPQLAVATRLQEYKDLIGASGSRYSDWLYTFFASKI 420
Query 421 EHVDRPKLLFSSSVMVNSQVVLNQAGQSGFEGGESAALGQMGGSISFNTVLGREQTYYFK 480
EHVDRPKLLFSSSVMVNSQVV+NQAGQSGF GGE+AALGQMGGSI+FNTVLGREQTYYFK
Sbjct 421 EHVDRPKLLFSSSVMVNSQVVMNQAGQSGFAGGEAAALGQMGGSIAFNTVLGREQTYYFK 480
Query 481 EPGYIFDMMTIRPVYFWTGIRPDYLEYRGPDYFNPIYNDIGYQDVPLWRLGYGWKADTVS 540
EPGYIFDM+TIRPVYFWTGIRPDYLEYRGPDYFNPIYNDIGYQDVPLWRLG+GWK+D VS
Sbjct 481 EPGYIFDMLTIRPVYFWTGIRPDYLEYRGPDYFNPIYNDIGYQDVPLWRLGFGWKSDAVS 540
Query 541 SLSVAKEPCYNEFRSSYDEVLGSLQATLTPKASTPLQSYWVQQRDFYLIGLSSNPNEVSP 600
S+SVAKEPCYNEFRSSYDEVLGSLQ+TLTPKAS PLQSYWVQQRDFYLIGLSS+ NE+SP
Sbjct 541 SVSVAKEPCYNEFRSSYDEVLGSLQSTLTPKASVPLQSYWVQQRDFYLIGLSSDLNEISP 600
Query 601 SMLFTNLNTVNNPFASDMEDNFFVNMSYKVVVKNLINKSFATRLSSR 647
SMLFTNL TVNNPF+SDMEDNFFVNMSYKVVVKNL+NKSFATRLSSR
Sbjct 601 SMLFTNLATVNNPFSSDMEDNFFVNMSYKVVVKNLVNKSFATRLSSR 647
> Alpavirinae_Human_feces_C_016_Microviridae_AG0274_putative.VP1
Length=641
Score = 70.1 bits (170), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 81/310 (26%), Positives = 121/310 (39%), Gaps = 65/310 (21%)
Query 327 GTGTDV------NIPSKVYFSATLNVPFLAAHPMAVCPSSPDRFSRLMPPG--------- 371
GTG ++ +P+ F A + P SPD F ++ G
Sbjct 282 GTGCELFKTFSDALPTSNSFVAMACAGIQGYGGLLSVPYSPDLFGNIIKQGSSPTAEIEV 341
Query 372 ----DSNSDVDFTGVKTIPQLAVATRLQEYKDLIGASGSRYSDWLYTFFASKIE--HVDR 425
DSN+D F+ +P+L + T++Q + D + SG R D T + +K +V++
Sbjct 342 IKALDSNTDTGFS--IAVPELRLKTKIQNWMDRLFISGGRVGDVFRTLWGTKSSAPYVNK 399
Query 426 PKLLFSSSVMVNSQVVLNQAGQSGFEGGESAALGQMGGSIS----FNTVLGREQTYYFKE 481
P L +N V +A +G GE A LGQ+ + F+ G + YY KE
Sbjct 400 PDFLGVWQASINPSNV--RAMANGSASGEDANLGQLAACVDRYCDFSDHSGID--YYAKE 455
Query 482 PG--YIFDMMTIRPVYFWTGIRPDYLEYRGPDYFNPIYNDIGYQDVPLWRL--------- 530
PG + M+ P Y G+ PD D FNP N IG+Q VP R
Sbjct 456 PGTFMLITMLVPEPAYS-QGLHPDLASISFGDDFNPELNGIGFQLVPRHRFSMMPRGFNL 514
Query 531 --------------GYGWKADTVSSLSVAKEPCYNEFRSSYDEVLGSLQATLTPKASTPL 576
G G D +++SV +E ++ R+ Y + G A
Sbjct 515 TGLDQQTSPWFGNAGTGVVIDP-NTVSVGEEVAWSWLRTDYSRLHGDF-------AQNGN 566
Query 577 QSYWVQQRDF 586
YWV R F
Sbjct 567 YQYWVLTRRF 576
Score = 23.9 bits (50), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 12/44 (27%), Positives = 22/44 (50%), Gaps = 0/44 (0%)
Query 26 SITPGIIYPVRIQFVNARDRVTLHQGIDIRSNPLGVPSFNPYVL 69
+I PG+ PV + +N +R+ ++S P+ P N + L
Sbjct 28 TIHPGLAIPVHHRHLNVGERIRGRIDELLQSQPMLGPLMNGFKL 71
> Pichovirinae_Pavin_723_Microviridae_AG0327_putative.VP1
Length=508
Score = 64.3 bits (155), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 54/188 (29%), Positives = 84/188 (45%), Gaps = 12/188 (6%)
Query 384 TIPQLAVATRLQEYKDLIGASGSRYSDWLYTFFA--SKIEHVDRPKLLFSSSVMVNSQVV 441
TI L A +LQE+ + G+RY + + T F S + + RP+ + V V
Sbjct 268 TINDLRRAFKLQEWLEKNARGGTRYIENILTHFGVRSSDKRLQRPEYITGVKSPVVVSEV 327
Query 442 LNQAGQSGFEGGESAALGQMGGSISFNTVLGREQTYYFKEPGYIFDMMTIRP-VYFWTGI 500
LN GQ G G M G + G+ +YY +E GYI +M++ P + GI
Sbjct 328 LNTTGQDG-----GLPQGNMAGH-GISVTSGKSGSYYCEEHGYIIGIMSVMPKTAYQQGI 381
Query 501 RPDYLEYRGPDYFNPIYNDIGYQDVPLWRLGYGWKADTVSSLSVAKEPCYNEFRSSYDEV 560
+L+ DYF P + +IG Q+V L Y + A+ + P Y E++ V
Sbjct 382 PRTFLKTDSLDYFWPTFANIGEQEVAKQEL-YAYTANANDTFGYV--PRYAEYKYMPSRV 438
Query 561 LGSLQATL 568
G + +L
Sbjct 439 AGEFRTSL 446
> Alpavirinae_Human_gut_30_040_Microviridae_AG0134_putative.VP1
Length=679
Score = 58.9 bits (141), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 51/191 (27%), Positives = 88/191 (46%), Gaps = 19/191 (10%)
Query 380 TGVKTIPQLAVATRLQEYKDLIGASGSRYSDWLYTFFASK-IEHVDRPKLLFSSSVMVNS 438
+G T+ L +A ++ + I S Y+ W+ T + S + H++ P L SS+ +
Sbjct 434 SGSFTLDTLNLAKKVYTMLNRIAVSDGSYNAWIQTVYTSGGLNHIETPLYLGGSSLEIEF 493
Query 439 QVVLNQAGQS----GFEGGESAALGQMGGSISFNTVLGREQTYYFKEPGYIFDMMTIRP- 493
Q V+N +G G G A GG+I F EPGYIF + +I P
Sbjct 494 QEVINNSGTEDQPLGALAGRGVATNHKGGNIVFKA----------DEPGYIFCITSITPR 543
Query 494 VYFWTGIRPD-YLEYRGPDYFNPIYNDIGYQDVPLWRLGYGWKADTVSSLSVAKEPCYNE 552
V ++ G D YLE D P + IG+QD L + + ++S+ K+P + +
Sbjct 544 VDYFQGNEWDLYLESL-DDLHKPQLDGIGFQDRLYRHLNSSCVREDL-NISIGKQPAWVQ 601
Query 553 FRSSYDEVLGS 563
+ ++ ++ G+
Sbjct 602 YMTNVNKTYGN 612
Score = 23.9 bits (50), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 18/91 (20%), Positives = 39/91 (43%), Gaps = 2/91 (2%)
Query 1 MGKQPFISHSVNGYSR--YDMPESKAFSITPGIIYPVRIQFVNARDRVTLHQGIDIRSNP 58
+G + +N Y+R +D ++ PG++ P + + D + ++P
Sbjct 10 LGGGKKMMTRLNNYNRSTHDNSFIMRTTMAPGVLVPTMTELLLPGDTYPIQIRCHTLTHP 69
Query 59 LGVPSFNPYVLRLHRFWVPLQLYHPELRVNS 89
P F + + F+ P++LY+ L N+
Sbjct 70 TIGPLFGSFKQQNDFFFCPIRLYNSMLHNNA 100
> Pichovirinae_JCVI_003_Microviridae_AG0344_putative.VP1
Length=505
Score = 57.8 bits (138), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 63/255 (25%), Positives = 110/255 (43%), Gaps = 30/255 (12%)
Query 384 TIPQLAVATRLQEYKDLIGASGSRYSDWLYTFF--ASKIEHVDRPKLLFSSSVMVNSQVV 441
TI L A +LQE+ +L+ GSRY + + TFF S + + RP+ + + V +
Sbjct 265 TITDLRRAFKLQEWLELLARGGSRYIEQIKTFFDVKSSDQRLQRPEYITGTKQPVIISEI 324
Query 442 LNQAGQS-GFEGGESAALGQMGGSISFNTVLGREQTYYFKEPGYIFDMMTIRP-VYFWTG 499
LN G++ G G + G G+ G YY +E G+I ++++ P + G
Sbjct 325 LNTTGETAGLPQGNMSGHGVAVGT-------GNVGKYYCEEHGFIIGILSVTPNTAYQQG 377
Query 500 IRPDYLEYRGPDYFNPIYNDIGYQDVPLWRLGYGWKADTVSSLSVAKEPCYNEFRSSYDE 559
I YL D++ P + IG Q + + + D + P Y E++
Sbjct 378 IPKHYLRTDKYDFYWPQFAHIGEQPIVNNEI---YAYDPTGDETFGYTPRYAEYKYMPSR 434
Query 560 VLGSLQATLTPKASTPLQSYWVQQRDFYLIGLSSNPNEVSPSMLFTNLNTVNNPFA-SDM 618
V G +++L W R F SS P +S + + + + FA +D
Sbjct 435 VAGEFRSSL---------DEWHAARIF-----SSLPT-LSQDFIEVDADDFDRIFAVTDG 479
Query 619 EDNFFVNMSYKVVVK 633
+DN ++++ KV +
Sbjct 480 DDNLYMHVLNKVKAR 494
Score = 25.0 bits (53), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 15/64 (23%), Positives = 28/64 (44%), Gaps = 0/64 (0%)
Query 14 YSRYDMPESKAFSITPGIIYPVRIQFVNARDRVTLHQGIDIRSNPLGVPSFNPYVLRLHR 73
Y+ +D+ S G + P+ D +TL ++ PL P + + + +H
Sbjct 17 YNTFDLTHDVKMSGRMGELLPIMNLECIPGDNITLGADSMVKFAPLLAPVMHRFNVTMHY 76
Query 74 FWVP 77
F+VP
Sbjct 77 FFVP 80
> Alpavirinae_Human_gut_32_012_Microviridae_AG0208_putative.VP1
Length=604
Score = 56.2 bits (134), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 63/249 (25%), Positives = 104/249 (42%), Gaps = 32/249 (13%)
Query 364 FSRLMPP------------GDSNSDVDFTG-VKTIP--QLAVATRLQEYKDLIGASGSRY 408
F R +PP DS VD K+I +A + +Q + DL A GSRY
Sbjct 261 FQRCLPPYYLESWLATSGYEDSEIKVDLGADGKSISFRNIAAQSHIQRWLDLALAGGSRY 320
Query 409 SDWLYTFF-ASKIEHVDRPKLLFSSSVMVNSQVVLNQAGQSGFEGGESAALGQMGGSISF 467
SD++ + F S+++H P L S + S V+ G A GQ G +F
Sbjct 321 SDYINSQFDVSRLKHTTSPLYLGSDRQYLGSNVIYQTTGAGDSASPLGAFAGQSSGGETF 380
Query 468 NTVLGREQTYYFKEPGYIFDMMTIRP-VYFWTGIRPDYLEYRGPDYFNPIYNDIGYQDVP 526
R ++Y+F E GY M ++ P V + G+ P E D + P ++I + +
Sbjct 381 -----RRRSYHFGENGYFVVMASLVPDVVYSRGMDPFNREKTLGDVYVPALDNIAMEPLM 435
Query 527 LWR------LGYGWKADTVSSLSVAKEPCYNEFRSSYDEVLGSLQATLT---PKASTPLQ 577
+ + L K D+V +LS+ + Y + ++ + +T L+
Sbjct 436 IEQVDAIPSLRSLQKVDSVYTLSIRPDKLIKNSALGYVPAWSKVMQNVSRAHGRLTTDLK 495
Query 578 SYWVQQRDF 586
YW+ RD+
Sbjct 496 -YWLLSRDY 503
Score = 37.0 bits (84), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 21/85 (25%), Positives = 42/85 (49%), Gaps = 0/85 (0%)
Query 6 FISHSVNGYSRYDMPESKAFSITPGIIYPVRIQFVNARDRVTLHQGIDIRSNPLGVPSFN 65
F++ S N S ++ +I PG + P+ + A D + +++ P+ P N
Sbjct 5 FLTRSRNKKSNVNLSHVSPTTIAPGNLVPISFTRIFAGDDLRFEPSAFVQAMPMNAPLVN 64
Query 66 PYVLRLHRFWVPLQLYHPELRVNSS 90
+ L L F+VP +LY+ L ++++
Sbjct 65 GFKLCLEYFFVPDRLYNWNLLMDNT 89
> Alpavirinae_Human_gut_31_126_Microviridae_AG0306_putative.VP1
Length=604
Score = 56.2 bits (134), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 63/249 (25%), Positives = 104/249 (42%), Gaps = 32/249 (13%)
Query 364 FSRLMPP------------GDSNSDVDFTG-VKTIP--QLAVATRLQEYKDLIGASGSRY 408
F R +PP DS VD K+I +A + +Q + DL A GSRY
Sbjct 261 FQRCLPPYYLESWLATSGYEDSEIKVDLGADGKSISFRNIAAQSHIQRWLDLALAGGSRY 320
Query 409 SDWLYTFF-ASKIEHVDRPKLLFSSSVMVNSQVVLNQAGQSGFEGGESAALGQMGGSISF 467
SD++ + F S+++H P L S + S V+ G A GQ G +F
Sbjct 321 SDYINSQFDVSRLKHTTSPLYLGSDRQYLGSNVIYQTTGAGDSASPLGAFAGQSSGGETF 380
Query 468 NTVLGREQTYYFKEPGYIFDMMTIRP-VYFWTGIRPDYLEYRGPDYFNPIYNDIGYQDVP 526
R ++Y+F E GY M ++ P V + G+ P E D + P ++I + +
Sbjct 381 -----RRRSYHFGENGYFVVMASLVPDVVYSRGMDPFNREKTLGDVYVPALDNIAMEPLM 435
Query 527 LWR------LGYGWKADTVSSLSVAKEPCYNEFRSSYDEVLGSLQATLT---PKASTPLQ 577
+ + L K D+V +LS+ + Y + ++ + +T L+
Sbjct 436 IEQVDAIPSLRSLQKVDSVYTLSIRPDKLIKNSALGYVPAWSKVMQNVSRAHGRLTTDLK 495
Query 578 SYWVQQRDF 586
YW+ RD+
Sbjct 496 -YWLLSRDY 503
Score = 37.0 bits (84), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 21/85 (25%), Positives = 42/85 (49%), Gaps = 0/85 (0%)
Query 6 FISHSVNGYSRYDMPESKAFSITPGIIYPVRIQFVNARDRVTLHQGIDIRSNPLGVPSFN 65
F++ S N S ++ +I PG + P+ + A D + +++ P+ P N
Sbjct 5 FLTRSRNKKSNVNLSHVSPTTIAPGNLVPISFTRIFAGDDLRFEPSAFVQAMPMNAPLVN 64
Query 66 PYVLRLHRFWVPLQLYHPELRVNSS 90
+ L L F+VP +LY+ L ++++
Sbjct 65 GFKLCLEYFFVPDRLYNWNLLMDNT 89
> Gokush_Human_feces_A_019_Microviridae_AG0438_putative.VP1
Length=569
Score = 54.7 bits (130), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 45/191 (24%), Positives = 92/191 (48%), Gaps = 15/191 (8%)
Query 384 TIPQLAVATRLQEYKDLIGASGSRYSDWLYTFFASKI--EHVDRPKLLFSSSVMVNSQVV 441
TI Q+ A ++Q+Y + + GSRY + +Y+ F ++I + V P+ L +M+N V
Sbjct 318 TINQIRQAFQVQKYFEELARGGSRYREQIYSLFRTRISDKTVQIPEYLGGDRIMINMSQV 377
Query 442 LNQAGQSGFE-GGESAALGQMG-GSISFNTVLGREQTYYFKEPGYIFDMMTIRPVY-FWT 498
+ +G + G +A+ G G +F T F+E G+I + +R + +
Sbjct 378 VQTSGTTDVSPQGNVSAMSVTGFGKSAF--------TKSFEEHGFIIGVCCVRHDHTYQQ 429
Query 499 GIRPDYLEYRGPDYFNPIYNDIGYQDVPLWRLGYGWKADTVSSLSVAKEPCYNEFRSSYD 558
G+ + DY+ P++ ++G Q V L + Y +T + + + C+ ++R +
Sbjct 430 GLERMFSRKNKLDYYFPVFANLGEQAV-LKKELYAQGTET-DNEAFGYQECWADYRMKPN 487
Query 559 EVLGSLQATLT 569
+ G+ ++ T
Sbjct 488 RICGAFRSNAT 498
> Pichovirinae_Pavin_279_Microviridae_AG0271_putative.VP1
Length=503
Score = 53.5 bits (127), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 68/260 (26%), Positives = 108/260 (42%), Gaps = 34/260 (13%)
Query 381 GVKTIPQLAVATRLQEYKDLIGASGSRYSDWLYTFFA--SKIEHVDRPKLLFSSSVMVNS 438
G TI L A RLQE+ + G+RY + + F S + RP+ + + V
Sbjct 260 GSTTINDLRRAMRLQEFLEKNARGGTRYIENIKMHFGVQSSDSRLQRPEYITGTKAPVVV 319
Query 439 QVVLNQAGQSGFEGGESAALGQMGGSISFNTVLGREQTYYFKEPGYIFDMMTIRPV-YFW 497
Q + S +G E G + S + G Y +E GYI +M+I P +
Sbjct 320 QEI-----TSTVKGIEEPQGNPTGKATSLQS--GHYGNYSVEEHGYIIGIMSIMPKPAYQ 372
Query 498 TGIRPDYLEYRGPDYFNPIYNDIGYQDVPLWRLGYGWKADTVSSLSVAKEPCYNEFRSSY 557
GI YL+ D++ P + +IG Q+V L YG+ + P Y E++
Sbjct 373 QGIPKSYLKTDVLDFYWPSFANIGEQEV-LQEEIYGYTPTPNAVFGYI--PRYAEYKYLQ 429
Query 558 DEVLGSLQATLTPKASTPLQSYWVQQRDFYLI-GLSSNPNEVSP---SMLFTNLNTVNNP 613
+ V G + TL +W R F I LS + EVSP + +F N
Sbjct 430 NRVCGLFRTTL---------DFWHLGRIFANIPTLSQSFIEVSPANQTRIFAN------- 473
Query 614 FASDMEDNFFVNMSYKVVVK 633
D +DN ++++ K+ +
Sbjct 474 -QEDYDDNIYIHVLNKMTAR 492
> Gokush_Human_gut_21_019_Microviridae_AG0380_putative.VP1
Length=563
Score = 53.9 bits (128), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 52/202 (26%), Positives = 90/202 (45%), Gaps = 15/202 (7%)
Query 372 DSNSDVDFTGVKTIPQLAVATRLQEYKDLIGASGSRYSDWLYTFFA--SKIEHVDRPKLL 429
DS D+D + + TI L A ++Q++ + + GSRY++ L +FF+ S + RP+ L
Sbjct 303 DSYVDLDTSSIFTINSLRTAFQMQKFYERLARGGSRYTEVLRSFFSVVSPDARLQRPEFL 362
Query 430 FSSSVMVNSQ-VVLNQAGQSGFEGGESAALGQMGGSISFNTVLGREQTYYFKEPGYIFDM 488
S + MVN + A S G +A G T F E GY+F
Sbjct 363 GSFTKMVNVNPIAQTSATDSTSPQGNLSAYGVTAAKF-------HGFTKSFVEHGYVFGF 415
Query 489 MTIRP-VYFWTGIRPDYLEYRGPDYFNPIYNDIGYQDVPLWRL-GYGWKADTVSSLSVAK 546
+ R + + GI +L D++ P + +G Q + L + G +ADT
Sbjct 416 VCARADLTYQQGINKMWLRSTVYDFYWPTFAHLGEQAIELREIYAQGSEADTT---VFGY 472
Query 547 EPCYNEFRSSYDEVLGSLQATL 568
+ Y E+R ++ G ++++
Sbjct 473 QERYAEYRYKPSQITGKFRSSV 494
Lambda K H a alpha
0.320 0.136 0.427 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 60102543