bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-37_CDS_annotation_glimmer3.pl_2_1
Length=238
Score E
Sequences producing significant alignments: (Bits) Value
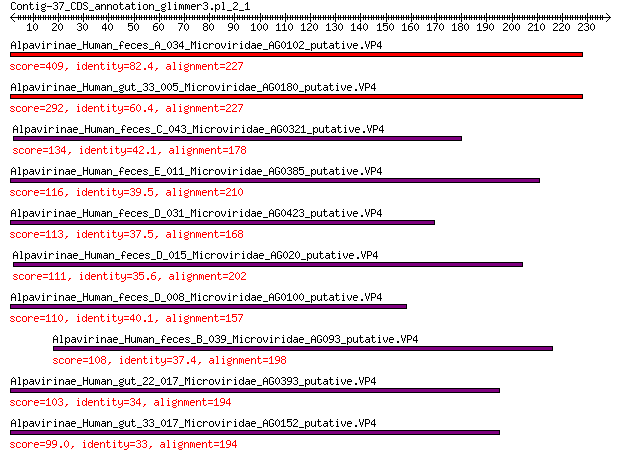
Alpavirinae_Human_feces_A_034_Microviridae_AG0102_putative.VP4 409 1e-142
Alpavirinae_Human_gut_33_005_Microviridae_AG0180_putative.VP4 292 5e-97
Alpavirinae_Human_feces_C_043_Microviridae_AG0321_putative.VP4 134 4e-38
Alpavirinae_Human_feces_E_011_Microviridae_AG0385_putative.VP4 116 3e-31
Alpavirinae_Human_feces_D_031_Microviridae_AG0423_putative.VP4 113 2e-30
Alpavirinae_Human_feces_D_015_Microviridae_AG020_putative.VP4 111 1e-29
Alpavirinae_Human_feces_D_008_Microviridae_AG0100_putative.VP4 110 2e-29
Alpavirinae_Human_feces_B_039_Microviridae_AG093_putative.VP4 108 8e-29
Alpavirinae_Human_gut_22_017_Microviridae_AG0393_putative.VP4 103 7e-27
Alpavirinae_Human_gut_33_017_Microviridae_AG0152_putative.VP4 99.0 3e-25
> Alpavirinae_Human_feces_A_034_Microviridae_AG0102_putative.VP4
Length=539
Score = 409 bits (1051), Expect = 1e-142, Method: Compositional matrix adjust.
Identities = 187/227 (82%), Positives = 216/227 (95%), Gaps = 0/227 (0%)
Query 1 MQLFFKRLNQNIRSVTNEKIYYYVVGEYGPTTFRPHFHILLFHDSKELRQSIRQFVSKSW 60
+QLFFKRLNQNIR +TNEKIYYYVVGEYGPTTFRPHFHILLFHDS++LR+SIRQFVSKSW
Sbjct 163 IQLFFKRLNQNIRRITNEKIYYYVVGEYGPTTFRPHFHILLFHDSRKLRESIRQFVSKSW 222
Query 61 RFGDTDTQPVWSSASCYVAGYVNSTACLPDFYKNFSHIKPFGRFSMHFAESAFNEVFKPQ 120
RFGD+DTQPVWSSASCYVAGYVNSTACLPDF+KN HIKPFGRFS++FAESAFNEVFKP+
Sbjct 223 RFGDSDTQPVWSSASCYVAGYVNSTACLPDFFKNSRHIKPFGRFSVNFAESAFNEVFKPE 282
Query 121 EDEEIFSLFYDGRMLELNGKPTLVRPKRSHINRLYPRLNKSKHASVDDDIRVATALSNIP 180
E+EEIFSLFYDGR+L LNGKPT+VRPKRSHINRLYPRL+KSKHA+V DDIR+A+ +S +P
Sbjct 283 ENEEIFSLFYDGRVLNLNGKPTIVRPKRSHINRLYPRLDKSKHATVVDDIRIASVVSRLP 342
Query 181 HVLAKFGFIDEVTDFEMSKRIYYLIRRYLEIDHTLKYAPEQLRLIYN 227
V+AKFGFIDEV+DFE+SKR+YYLIRR+LE+D +L YA ++LR+IYN
Sbjct 343 QVVAKFGFIDEVSDFELSKRVYYLIRRHLEVDCSLDYASDELRVIYN 389
> Alpavirinae_Human_gut_33_005_Microviridae_AG0180_putative.VP4
Length=536
Score = 292 bits (747), Expect = 5e-97, Method: Compositional matrix adjust.
Identities = 137/227 (60%), Positives = 172/227 (76%), Gaps = 0/227 (0%)
Query 1 MQLFFKRLNQNIRSVTNEKIYYYVVGEYGPTTFRPHFHILLFHDSKELRQSIRQFVSKSW 60
+QLF KR+NQ I+ T+EKIY Y VGEYGP TFRPHFH+L F DS+ L Q Q V K+W
Sbjct 162 VQLFHKRINQQIKKYTDEKIYSYTVGEYGPKTFRPHFHLLFFFDSERLAQVFGQLVDKAW 221
Query 61 RFGDTDTQPVWSSASCYVAGYVNSTACLPDFYKNFSHIKPFGRFSMHFAESAFNEVFKPQ 120
RFG++DTQ VWSSAS YVAGY+NS+ CLP+FY+ I PFGRFS +FAE F E FKP+
Sbjct 222 RFGNSDTQRVWSSASSYVAGYLNSSHCLPEFYRCNRKIAPFGRFSQYFAERPFIEAFKPE 281
Query 121 EDEEIFSLFYDGRMLELNGKPTLVRPKRSHINRLYPRLNKSKHASVDDDIRVATALSNIP 180
E+E++F F +G L L GKPTL RPKRS INRLYP L++S + VD ++R A +S IP
Sbjct 282 ENEKVFDKFVNGIYLSLGGKPTLCRPKRSLINRLYPVLDRSAVSDVDSNVRTALFVSKIP 341
Query 181 HVLAKFGFIDEVTDFEMSKRIYYLIRRYLEIDHTLKYAPEQLRLIYN 227
VLAKFGF+DEVT FE +KR +++I++YL++DH+L APE LR IYN
Sbjct 342 QVLAKFGFLDEVTLFEQAKRTFFVIKKYLQVDHSLDNAPETLRFIYN 388
> Alpavirinae_Human_feces_C_043_Microviridae_AG0321_putative.VP4
Length=546
Score = 134 bits (338), Expect = 4e-38, Method: Compositional matrix adjust.
Identities = 75/184 (41%), Positives = 102/184 (55%), Gaps = 6/184 (3%)
Query 2 QLFFKRLNQNIRSVTNEKIYYYVVGEYGPTTFRPHFHILLFHDSKELRQSIRQFVSKSWR 61
QLF KR+ + IR+ T+EKI+ Y+VGEY P FRPHFHIL F +S EL QS V W+
Sbjct 161 QLFMKRVRKQIRNYTDEKIHTYIVGEYSPKHFRPHFHILFFFNSNELSQSFGSIVRSCWK 220
Query 62 FGDTDTQPVWSSASCYVAGYVNSTACLPDFYKNFSHIKPFGRFSMHFAESAFNEVFKPQE 121
FG D A YVAGYVNS A LP + I PFGRFS HF+ES F++ +
Sbjct 221 FGLVDWSQSRGDAESYVAGYVNSFARLPYHLGSSPKIAPFGRFSNHFSESCFDDAKQTLR 280
Query 122 DE------EIFSLFYDGRMLELNGKPTLVRPKRSHINRLYPRLNKSKHASVDDDIRVATA 175
D +F+ F +G +NGK L RP RS ++ + R + S + + + A
Sbjct 281 DSFFGQKTPVFAPFLNGVTNLVNGKLLLTRPSRSCVDSCFFRKARDGRLSSHELLHLIRA 340
Query 176 LSNI 179
+ ++
Sbjct 341 VCHV 344
> Alpavirinae_Human_feces_E_011_Microviridae_AG0385_putative.VP4
Length=611
Score = 116 bits (290), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 83/222 (37%), Positives = 120/222 (54%), Gaps = 19/222 (9%)
Query 1 MQLFFKRLNQNIRS--VTNEKIYYYVVGEYGPTTFRPHFHILLFHDSKELRQSIRQFVSK 58
++LFFKRL +N+ S T+ KI YYVV EYGP T+RPH+H LLF +S+E+ Q++R+ +SK
Sbjct 209 IELFFKRLRRNLDSHGFTSSKICYYVVSEYGPQTYRPHWHCLLFFNSEEITQTLREDISK 268
Query 59 SWRFGDTDTQPVWSSASCYVAGYVNSTACLPDFYKNFSHIKPFGRFSMHFAESAFNEVFK 118
+W +G D +A+ YVA YVNS ACLP Y I+P S F N+VF
Sbjct 269 AWSYGRIDYSLSRGAAASYVAAYVNSAACLPFLYVGQKEIRPRSFHSKGFGS---NKVFP 325
Query 119 PQED----EEIFSLFYDGRMLELNGKPTLVRPKRSHINRLYPRLNKSKHASVDDDIRVAT 174
D +I LF+DG ++ NGK +RP RS ++PR + + D ++
Sbjct 326 KSSDVSEISKISDLFFDGVNVDSNGKVVNIRPVRSCELAVFPRFSNDFFSDSDTCCKLFQ 385
Query 175 ALSNIPHVLAKFGFIDEVT------DFEMSKRIYYLIRRYLE 210
++ P L G++ T DF +S L+R Y E
Sbjct 386 SVIETPERLVSRGYLGIDTPNFGSDDFRLSD----LVRAYSE 423
> Alpavirinae_Human_feces_D_031_Microviridae_AG0423_putative.VP4
Length=532
Score = 113 bits (282), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 63/169 (37%), Positives = 92/169 (54%), Gaps = 2/169 (1%)
Query 1 MQLFFKRLNQNIRSV-TNEKIYYYVVGEYGPTTFRPHFHILLFHDSKELRQSIRQFVSKS 59
+ L+ KR+ + I + NE I+ Y+VGEYGP++FRPHFH+LLF DS EL Q+I + S
Sbjct 165 VSLYMKRVRKYISKLGINETIHTYIVGEYGPSSFRPHFHLLLFFDSDELAQNIIRIASSC 224
Query 60 WRFGDTDTQPVWSSASCYVAGYVNSTACLPDFYKNFSHIKPFGRFSMHFAESAFNEVFKP 119
WRFG D A YV+ Y+NS + +P + I+PF RFS F S F K
Sbjct 225 WRFGRVDCSASRGDAEDYVSSYLNSFSSIPLHIQEIRAIRPFARFSNKFGYSFFESSIKK 284
Query 120 QEDEEIFSLFYDGRMLELNGKPTLVRPKRSHINRLYPRLNKSKHASVDD 168
+ F +G+ L NG T + P R+ I+ + R +H+ + +
Sbjct 285 AQSGN-FDEILNGKSLPYNGFDTTIFPWRAIIDTCFYRPALRRHSDIHE 332
> Alpavirinae_Human_feces_D_015_Microviridae_AG020_putative.VP4
Length=563
Score = 111 bits (277), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 72/204 (35%), Positives = 102/204 (50%), Gaps = 4/204 (2%)
Query 2 QLFFKRLNQNIRSVTN--EKIYYYVVGEYGPTTFRPHFHILLFHDSKELRQSIRQFVSKS 59
QLF KRL + + EKI+ YVV EY P TFRPHFHIL F DS E+ ++ RQ V +S
Sbjct 216 QLFAKRLRKYLSKKVGKYEKIHSYVVSEYSPKTFRPHFHILFFFDSDEIAKNFRQAVYQS 275
Query 60 WRFGDTDTQPVWSSASCYVAGYVNSTACLPDFYKNFSHIKPFGRFSMHFAESAFNEVFKP 119
WR G DTQ A+ YV+ Y+NS +P YK I+P RFS F E +
Sbjct 276 WRLGRVDTQLAREQANSYVSNYLNSVVSIPFVYKAKKSIRPRSRFSNLFGFEEVKEGIRK 335
Query 120 QEDEEIFSLFYDGRMLELNGKPTLVRPKRSHINRLYPRLNKSKHASVDDDIRVATALSNI 179
+D+ + +DG N K P S I+RL+PR + + ++ + I
Sbjct 336 AQDKR--AALFDGLSYISNQKFVRYVPSGSLIDRLFPRFTYYDGSFLRRSSQIYGVVQQI 393
Query 180 PHVLAKFGFIDEVTDFEMSKRIYY 203
+ A+ +E T +S+ I +
Sbjct 394 LRLFARNEPFEEATPGNVSEFICW 417
> Alpavirinae_Human_feces_D_008_Microviridae_AG0100_putative.VP4
Length=532
Score = 110 bits (274), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 63/158 (40%), Positives = 86/158 (54%), Gaps = 2/158 (1%)
Query 1 MQLFFKRLNQNIRSV-TNEKIYYYVVGEYGPTTFRPHFHILLFHDSKELRQSIRQFVSKS 59
+ L+ KR+ + I + NE I+ YVVGEYGP TFRPHFH+LLF +S EL QSI +
Sbjct 165 VSLYMKRVRKYISKLGINETIHTYVVGEYGPVTFRPHFHLLLFFNSDELAQSIVRIARSC 224
Query 60 WRFGDTDTQPVWSSASCYVAGYVNSTACLPDFYKNFSHIKPFGRFSMHFAESAFNEVFKP 119
WRFG D A YV+ Y+NS + +P + I+PF RFS F S F K
Sbjct 225 WRFGRVDCSASRGDAEDYVSSYLNSFSSIPLHIQEIRAIRPFARFSNKFGFSFFESSIKK 284
Query 120 QEDEEIFSLFYDGRMLELNGKPTLVRPKRSHINRLYPR 157
+ + F +G+ L NG T + P R+ I+ + R
Sbjct 285 AQSGD-FDEILNGKSLPYNGFNTTIFPWRAIIDTCFYR 321
> Alpavirinae_Human_feces_B_039_Microviridae_AG093_putative.VP4
Length=559
Score = 108 bits (271), Expect = 8e-29, Method: Compositional matrix adjust.
Identities = 74/206 (36%), Positives = 110/206 (53%), Gaps = 15/206 (7%)
Query 18 EKIYYYVVGEYGPTTFRPHFHILLFHDSKELRQSIRQFVSKSWRFGDTDTQPVWSSASCY 77
E ++Y VGEYGP FRPH+H+LLF +S ++ + +R KSW+ G +D Q A Y
Sbjct 207 ETFHFYAVGEYGPVHFRPHYHLLLFTNSDKVSEVLRYCHDKSWKLGRSDFQRSAGGAGSY 266
Query 78 VAGYVNSTACLPDFYKNFSHIKPFGRFSMHFAESA--FNEVFKP--QEDEEIFSLFYDGR 133
VA YVNS P Y++ +P R S+ F E F E P Q +++I S+ +GR
Sbjct 267 VASYVNSLCSAPLLYRSCRAFRPKSRASVGFFEKGCDFVEDDDPYAQIEQKIDSVV-NGR 325
Query 134 MLELNGKPTLVRPKRSHINRLYPRLNKSKHASVDDDIRVAT---ALSNIPHVLAKFGFID 190
+ NG P S+I L PR + +++ DD I +A A+ + P +A+FGF D
Sbjct 326 VYNFNGVSVRSTPPLSYIRTLLPRFSSARN---DDGIAIARILYAVHSTPKRIARFGFAD 382
Query 191 EVTDFEMS-KRIYYLIRRYLEIDHTL 215
D +S R YY +YL+++ L
Sbjct 383 YKQDSILSLVRTYY---QYLKVNPIL 405
> Alpavirinae_Human_gut_22_017_Microviridae_AG0393_putative.VP4
Length=562
Score = 103 bits (257), Expect = 7e-27, Method: Compositional matrix adjust.
Identities = 66/196 (34%), Positives = 100/196 (51%), Gaps = 5/196 (3%)
Query 1 MQLFFKRLNQNIRSVTNEKIYYYVVGEYGPTTFRPHFHILLFHDSKELRQSIRQFVSKSW 60
+ LF KRL +EK+ YY V EYGPT+FRPH+H+LLF +S+ +++ + VSK+W
Sbjct 171 LDLFLKRLRS---YYLDEKLRYYAVSEYGPTSFRPHWHLLLFSNSERFSRTVCENVSKAW 227
Query 61 RFGDTDTQPVWSSASCYVAGYVNSTACLPDFYKNFSH-IKPFGRFSMHFAES-AFNEVFK 118
+G D A+ YVA YVNS LPDFY ++P S+ F ES F +
Sbjct 228 SYGRCDASLSRGFAAPYVASYVNSFVALPDFYTQMPKVVRPKSFHSIGFTESNLFPRKVR 287
Query 119 PQEDEEIFSLFYDGRMLELNGKPTLVRPKRSHINRLYPRLNKSKHASVDDDIRVATALSN 178
E +EI +G +E +G ++P ++ RL+PR + + S ++ A
Sbjct 288 VAEVDEITDKCLNGVRVERDGYFRTIKPTWPYLLRLFPRFSDAIRKSPSSIYQLLFAAFT 347
Query 179 IPHVLAKFGFIDEVTD 194
P + + G D D
Sbjct 348 APERVIRSGCADIGCD 363
> Alpavirinae_Human_gut_33_017_Microviridae_AG0152_putative.VP4
Length=565
Score = 99.0 bits (245), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 64/196 (33%), Positives = 100/196 (51%), Gaps = 5/196 (3%)
Query 1 MQLFFKRLNQNIRSVTNEKIYYYVVGEYGPTTFRPHFHILLFHDSKELRQSIRQFVSKSW 60
+ LF KRL EK+ YY V EYGPT++RPH+H+LLF +S++ ++I + VSK+W
Sbjct 171 LDLFLKRLRS---YYPYEKLRYYAVSEYGPTSYRPHWHLLLFSNSEQFSKTILENVSKAW 227
Query 61 RFGDTDTQPVWSSASCYVAGYVNSTACLPDFYKNFSH-IKPFGRFSMHFAES-AFNEVFK 118
+G D A+ YVA YVNS LP FY ++P S+ F ES F +
Sbjct 228 SYGRCDASLSRGFAAPYVASYVNSFVALPSFYTEMPRFLRPKSFHSIGFTESNLFPRKVR 287
Query 119 PQEDEEIFSLFYDGRMLELNGKPTLVRPKRSHINRLYPRLNKSKHASVDDDIRVATALSN 178
E +E+ +G +E +G +++P ++ RL+PR + + S ++ A
Sbjct 288 ISEIDEVTDKCLNGVRVERDGYFRILKPSWPYLLRLFPRFSDAIRKSPSSIYQLLFAAFT 347
Query 179 IPHVLAKFGFIDEVTD 194
P + + G D D
Sbjct 348 APARVIRSGCADIGCD 363
Lambda K H a alpha
0.326 0.140 0.429 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 18832452