bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-37_CDS_annotation_glimmer3.pl_2_2
Length=147
Score E
Sequences producing significant alignments: (Bits) Value
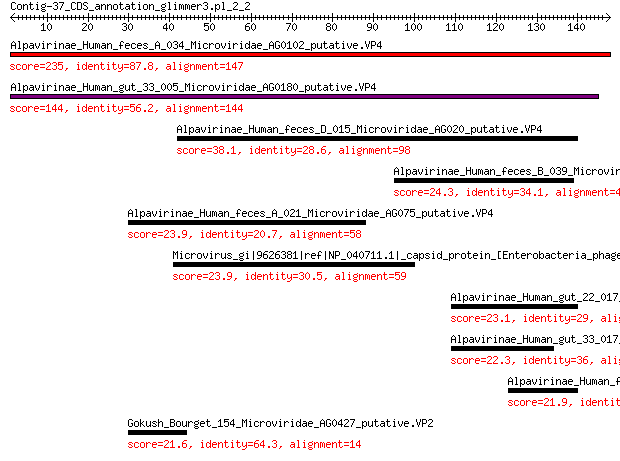
Alpavirinae_Human_feces_A_034_Microviridae_AG0102_putative.VP4 235 9e-77
Alpavirinae_Human_gut_33_005_Microviridae_AG0180_putative.VP4 144 6e-43
Alpavirinae_Human_feces_D_015_Microviridae_AG020_putative.VP4 38.1 1e-05
Alpavirinae_Human_feces_B_039_Microviridae_AG093_putative.VP4 24.3 0.53
Alpavirinae_Human_feces_A_021_Microviridae_AG075_putative.VP4 23.9 0.65
Microvirus_gi|9626381|ref|NP_040711.1|_capsid_protein_[Enteroba... 23.9 0.76
Alpavirinae_Human_gut_22_017_Microviridae_AG0393_putative.VP4 23.1 1.2
Alpavirinae_Human_gut_33_017_Microviridae_AG0152_putative.VP4 22.3 2.0
Alpavirinae_Human_feces_E_011_Microviridae_AG0385_putative.VP4 21.9 3.2
Gokush_Bourget_154_Microviridae_AG0427_putative.VP2 21.6 3.7
> Alpavirinae_Human_feces_A_034_Microviridae_AG0102_putative.VP4
Length=539
Score = 235 bits (600), Expect = 9e-77, Method: Compositional matrix adjust.
Identities = 129/147 (88%), Positives = 136/147 (93%), Gaps = 0/147 (0%)
Query 1 LSLYINFSDESGCSAvyrlllnyrnlvnnWITAPVGSVAFTGQLRYAVRSIHSFYDYCAK 60
LSLYINFSDESGC+A+YRLLL YRNLV NWITAPVGSVAFTGQL AVR+IHSFYDYC++
Sbjct 393 LSLYINFSDESGCAAIYRLLLQYRNLVKNWITAPVGSVAFTGQLHRAVRAIHSFYDYCSR 452
Query 61 RSLRDQLLKVEKWSNDSYVRENLSIYYFYPLTDVDIMKSSYSEIVFYSPVLRASYADYAA 120
RSL DQL+KVEKWSNDSYVR NLSIYYFYPLTDVDIM S+SEIVF S VLRASYADYAA
Sbjct 453 RSLHDQLVKVEKWSNDSYVRANLSIYYFYPLTDVDIMMKSFSEIVFDSQVLRASYADYAA 512
Query 121 DNRERIKHKALNDKNSLLIAAVDKKPV 147
DNRERIKHKALNDKNSLLIA VDKKPV
Sbjct 513 DNRERIKHKALNDKNSLLIAEVDKKPV 539
> Alpavirinae_Human_gut_33_005_Microviridae_AG0180_putative.VP4
Length=536
Score = 144 bits (364), Expect = 6e-43, Method: Compositional matrix adjust.
Identities = 81/144 (56%), Positives = 104/144 (72%), Gaps = 0/144 (0%)
Query 1 LSLYINFSDESGCSAvyrlllnyrnlvnnWITAPVGSVAFTGQLRYAVRSIHSFYDYCAK 60
LSLY+NFS+ GCSAVYRL L YRNL ++WIT P +AF GQLR A R+I++FY Y
Sbjct 392 LSLYLNFSELEGCSAVYRLFLAYRNLCDHWITVPSNHIAFYGQLRKAFRTIYAFYSYMDS 451
Query 61 RSLRDQLLKVEKWSNDSYVRENLSIYYFYPLTDVDIMKSSYSEIVFYSPVLRASYADYAA 120
+ L DQL+KV+ WS ++Y+ + YFYPLTD ++M+SSY E++ S RA+YA+ AA
Sbjct 452 KYLHDQLVKVQSWSQNNYLSTQVDFRYFYPLTDGELMRSSYREVLALSSFCRAAYAEVAA 511
Query 121 DNRERIKHKALNDKNSLLIAAVDK 144
DNR+RIKHK LND N LIA DK
Sbjct 512 DNRQRIKHKYLNDLNCALIAECDK 535
> Alpavirinae_Human_feces_D_015_Microviridae_AG020_putative.VP4
Length=563
Score = 38.1 bits (87), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 28/99 (28%), Positives = 52/99 (53%), Gaps = 11/99 (11%)
Query 42 GQLRYAVRSIHSFYDYCAKRSLRDQL-LKVEKWSNDSYVRENLSIYYFYPLTDVDIMKSS 100
G R ++++ FYDY +SL++QL ++ +S Y E L +Y P ++ ++K+
Sbjct 475 GSFRSKLKAVELFYDYRDYQSLKNQLYMQQLVFSELGYSDELLDSFYVKP--NLKVLKNI 532
Query 101 YSEIVFYSPVLRASYADYAADNRERIKHKALNDKNSLLI 139
Y+E + +Y + R+KHK LND+N + +
Sbjct 533 YTE--------KWHDTNYHEVHYFRVKHKVLNDQNDVFL 563
> Alpavirinae_Human_feces_B_039_Microviridae_AG093_putative.VP4
Length=559
Score = 24.3 bits (51), Expect = 0.53, Method: Compositional matrix adjust.
Identities = 15/44 (34%), Positives = 21/44 (48%), Gaps = 3/44 (7%)
Query 95 DIMKSSYSEIVFYSPVLRASYADYAADNRERIKHKALNDKNSLL 138
D++ E V LR A + R+ IKHK LND N++
Sbjct 515 DVLSDVSCETVQLLEQLRLRSATFC---RDMIKHKRLNDANNIF 555
> Alpavirinae_Human_feces_A_021_Microviridae_AG075_putative.VP4
Length=547
Score = 23.9 bits (50), Expect = 0.65, Method: Compositional matrix adjust.
Identities = 12/61 (20%), Positives = 30/61 (49%), Gaps = 3/61 (5%)
Query 30 WITAPVGSVAFTGQLRYAVRSIHSFYDYCAKRSLRDQLLKVEKWSNDSYVR---ENLSIY 86
W + + F ++ A+R+++ +Y R L +Q + ++ + + R ++L +Y
Sbjct 424 WTSCDICGFGFDREIELAIRNVNRYYSETEYRYLVNQYEDILRFIDSEFYRGEVDDLLMY 483
Query 87 Y 87
Y
Sbjct 484 Y 484
> Microvirus_gi|9626381|ref|NP_040711.1|_capsid_protein_[Enterobacteria_phage_phiX174_sensu_lato]
Length=427
Score = 23.9 bits (50), Expect = 0.76, Method: Compositional matrix adjust.
Identities = 18/59 (31%), Positives = 29/59 (49%), Gaps = 8/59 (14%)
Query 41 TGQLRYAVRSIHSFYDYCAKRSLRDQLLKVEKWSNDSYVRENLSIYYFYPLTDVDIMKS 99
+G L+ V H YD C Q +++ +W +S V+ N+++Y P T IM S
Sbjct 377 SGDLQERVLIRHHDYDQCF------QSVQLLQW--NSQVKFNVTVYRNLPTTRDSIMTS 427
> Alpavirinae_Human_gut_22_017_Microviridae_AG0393_putative.VP4
Length=562
Score = 23.1 bits (48), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 9/31 (29%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 109 PVLRASYADYAADNRERIKHKALNDKNSLLI 139
P++R A R+++KHK +ND + + +
Sbjct 532 PLVRRLAAASLMKCRDKVKHKKVNDLSGIFL 562
> Alpavirinae_Human_gut_33_017_Microviridae_AG0152_putative.VP4
Length=565
Score = 22.3 bits (46), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 9/25 (36%), Positives = 15/25 (60%), Gaps = 0/25 (0%)
Query 109 PVLRASYADYAADNRERIKHKALND 133
P++R A R+++KHK +ND
Sbjct 532 PLVRRLAAASLMKCRDKVKHKKIND 556
> Alpavirinae_Human_feces_E_011_Microviridae_AG0385_putative.VP4
Length=611
Score = 21.9 bits (45), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 6/17 (35%), Positives = 12/17 (71%), Gaps = 0/17 (0%)
Query 123 RERIKHKALNDKNSLLI 139
++++KHK ND +L+
Sbjct 592 KDKVKHKEFNDMQGVLL 608
> Gokush_Bourget_154_Microviridae_AG0427_putative.VP2
Length=253
Score = 21.6 bits (44), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 9/19 (47%), Positives = 12/19 (63%), Gaps = 5/19 (26%)
Query 30 WITAPVGSVA-----FTGQ 43
W+T+P+ SVA F GQ
Sbjct 6 WLTSPIASVASGVAGFLGQ 24
Lambda K H a alpha
0.320 0.133 0.392 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 9663476