bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-4_CDS_annotation_glimmer3.pl_2_1
Length=155
Score E
Sequences producing significant alignments: (Bits) Value
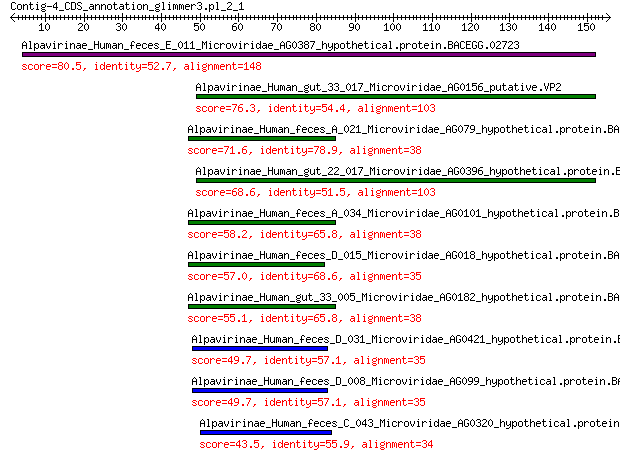
Alpavirinae_Human_feces_E_011_Microviridae_AG0387_hypothetical.... 80.5 4e-20
Alpavirinae_Human_gut_33_017_Microviridae_AG0156_putative.VP2 76.3 1e-18
Alpavirinae_Human_feces_A_021_Microviridae_AG079_hypothetical.p... 71.6 5e-17
Alpavirinae_Human_gut_22_017_Microviridae_AG0396_hypothetical.p... 68.6 7e-16
Alpavirinae_Human_feces_A_034_Microviridae_AG0101_hypothetical.... 58.2 2e-12
Alpavirinae_Human_feces_D_015_Microviridae_AG018_hypothetical.p... 57.0 6e-12
Alpavirinae_Human_gut_33_005_Microviridae_AG0182_hypothetical.p... 55.1 2e-11
Alpavirinae_Human_feces_D_031_Microviridae_AG0421_hypothetical.... 49.7 1e-09
Alpavirinae_Human_feces_D_008_Microviridae_AG099_hypothetical.p... 49.7 1e-09
Alpavirinae_Human_feces_C_043_Microviridae_AG0320_hypothetical.... 43.5 2e-07
> Alpavirinae_Human_feces_E_011_Microviridae_AG0387_hypothetical.protein.BACEGG.02723
Length=410
Score = 80.5 bits (197), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 78/151 (52%), Positives = 103/151 (68%), Gaps = 3/151 (2%)
Query 4 LDFIPVVG---DIASSIGNVVSTnkannanmavnrmnnefnaaeaeKARQFQLDMWNKTN 60
LD + +G S +GN+ + +N+ NM +NRMNNEFNA EAEKARQ+Q +MWNKTN
Sbjct 47 LDPLSAIGVGLGAVSGVGNIFGSALSNSQNMKINRMNNEFNAREAEKARQYQSEMWNKTN 106
Query 61 EYNSASAQRSRLEEAGLNPYLMMNggsagtaqssgsstpasassplsMQRQDFSGLSNTL 120
++NS R RL+EAG NPYL ++ + GTAQS+GSS+PASA+ P+ F G N L
Sbjct 107 DWNSPKNVRKRLQEAGYNPYLGLDSSNVGTAQSAGSSSPASAAPPIQNNPIQFDGFQNAL 166
Query 121 ASALQISNQTKETNANVQTLQSQKSLYDAQA 151
++A+Q+SN TK +NA LQ QK L DAQA
Sbjct 167 STAIQMSNSTKVSNAEANNLQGQKGLADAQA 197
> Alpavirinae_Human_gut_33_017_Microviridae_AG0156_putative.VP2
Length=353
Score = 76.3 bits (186), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 56/105 (53%), Positives = 72/105 (69%), Gaps = 6/105 (6%)
Query 49 RQFQLDMWNKTNEYNSASAQRSRLEEAGLNPYLMMNggsagtaqssgsstpasassplsM 108
R FQ +MWNK N YN+ASAQR RLEEAGLNPYLMMNGGS+G +QS+G+ AS+S
Sbjct 54 RDFQENMWNKENTYNTASAQRQRLEEAGLNPYLMMNGGSSGVSQSAGTGASASSSGTAVF 113
Query 109 Q--RQDFSGLSNTLASALQISNQTKETNANVQTLQSQKSLYDAQA 151
Q + DFSG+ + S Q +Q ++ A V +Q Q++L DAQA
Sbjct 114 QPFQADFSGIQQAIGSVFQ--SQVRQ--AQVSQMQGQRNLADAQA 154
> Alpavirinae_Human_feces_A_021_Microviridae_AG079_hypothetical.protein.BACEGG.02723
Length=397
Score = 71.6 bits (174), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 30/38 (79%), Positives = 35/38 (92%), Gaps = 0/38 (0%)
Query 47 KARQFQLDMWNKTNEYNSASAQRSRLEEAGLNPYLMMN 84
K R F++DMWNKTNEYNSA+ QR+RLEEAGLNPY+MMN
Sbjct 26 KNRDFEVDMWNKTNEYNSATNQRARLEEAGLNPYMMMN 63
> Alpavirinae_Human_gut_22_017_Microviridae_AG0396_hypothetical.protein.BACPLE
Length=386
Score = 68.6 bits (166), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 53/105 (50%), Positives = 70/105 (67%), Gaps = 6/105 (6%)
Query 49 RQFQLDMWNKTNEYNSASAQRSRLEEAGLNPYLMMNggsagtaqssgsstpasassplsM 108
R +Q +MWNK N YN+ASAQR RLEEAGLNPYLMMNGGSAG AQS+G+ + AS+S M
Sbjct 54 RNWQENMWNKENAYNTASAQRQRLEEAGLNPYLMMNGGSAGVAQSAGTGSAASSSGNAVM 113
Query 109 Q--RQDFSGLSNTLASALQISNQTKETNANVQTLQSQKSLYDAQA 151
Q + D+SG+ +++ + Q E + LQ + L DA+A
Sbjct 114 QPFQADYSGIGSSIGNIFQYELMQSEKS----QLQGARQLADAKA 154
> Alpavirinae_Human_feces_A_034_Microviridae_AG0101_hypothetical.protein.BACPLE
Length=365
Score = 58.2 bits (139), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 25/38 (66%), Positives = 32/38 (84%), Gaps = 0/38 (0%)
Query 47 KARQFQLDMWNKTNEYNSASAQRSRLEEAGLNPYLMMN 84
K+R F M++ +NE+NSA +QR+RLEEAGLNPYLMMN
Sbjct 50 KSRDFAKSMFDASNEWNSAKSQRARLEEAGLNPYLMMN 87
> Alpavirinae_Human_feces_D_015_Microviridae_AG018_hypothetical.protein.BACEGG.02723
Length=427
Score = 57.0 bits (136), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 24/35 (69%), Positives = 28/35 (80%), Gaps = 0/35 (0%)
Query 47 KARQFQLDMWNKTNEYNSASAQRSRLEEAGLNPYL 81
KAR FQLDMWNK N YN+ +AQR+RLEE G N Y+
Sbjct 79 KARAFQLDMWNKENAYNTPAAQRARLEEGGYNAYM 113
> Alpavirinae_Human_gut_33_005_Microviridae_AG0182_hypothetical.protein.BACEGG.02723
Length=354
Score = 55.1 bits (131), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 25/38 (66%), Positives = 30/38 (79%), Gaps = 0/38 (0%)
Query 47 KARQFQLDMWNKTNEYNSASAQRSRLEEAGLNPYLMMN 84
K+R F M++ TNE+NSA QR+RLE AGLNPYLMMN
Sbjct 50 KSRDFAKSMFDATNEWNSAKNQRARLEAAGLNPYLMMN 87
> Alpavirinae_Human_feces_D_031_Microviridae_AG0421_hypothetical.protein.BACEGG.02723
Length=418
Score = 49.7 bits (117), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 20/35 (57%), Positives = 26/35 (74%), Gaps = 0/35 (0%)
Query 48 ARQFQLDMWNKTNEYNSASAQRSRLEEAGLNPYLM 82
++Q+ LD WN+ N YN SAQR+R+E AG NPY M
Sbjct 67 SQQWNLDQWNRENAYNDPSAQRARMEAAGFNPYNM 101
> Alpavirinae_Human_feces_D_008_Microviridae_AG099_hypothetical.protein.BACEGG.02723
Length=418
Score = 49.7 bits (117), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 20/35 (57%), Positives = 26/35 (74%), Gaps = 0/35 (0%)
Query 48 ARQFQLDMWNKTNEYNSASAQRSRLEEAGLNPYLM 82
++Q+ LD WN+ N YN SAQR+R+E AG NPY M
Sbjct 67 SQQWNLDQWNRENAYNDPSAQRARMEAAGFNPYNM 101
> Alpavirinae_Human_feces_C_043_Microviridae_AG0320_hypothetical.protein.BACEGG.02723
Length=396
Score = 43.5 bits (101), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 19/34 (56%), Positives = 24/34 (71%), Gaps = 0/34 (0%)
Query 50 QFQLDMWNKTNEYNSASAQRSRLEEAGLNPYLMM 83
++ L WN+ N YN+ +AQRSRLE AGLN L M
Sbjct 69 EWNLQQWNRENAYNTPAAQRSRLEAAGLNAALAM 102
Lambda K H a alpha
0.312 0.124 0.331 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 10396563