bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-4_CDS_annotation_glimmer3.pl_2_8
Length=138
Score E
Sequences producing significant alignments: (Bits) Value
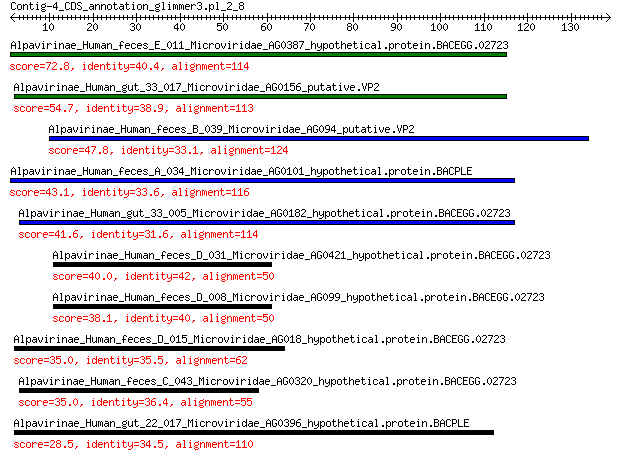
Alpavirinae_Human_feces_E_011_Microviridae_AG0387_hypothetical.... 72.8 2e-17
Alpavirinae_Human_gut_33_017_Microviridae_AG0156_putative.VP2 54.7 2e-11
Alpavirinae_Human_feces_B_039_Microviridae_AG094_putative.VP2 47.8 4e-09
Alpavirinae_Human_feces_A_034_Microviridae_AG0101_hypothetical.... 43.1 2e-07
Alpavirinae_Human_gut_33_005_Microviridae_AG0182_hypothetical.p... 41.6 5e-07
Alpavirinae_Human_feces_D_031_Microviridae_AG0421_hypothetical.... 40.0 2e-06
Alpavirinae_Human_feces_D_008_Microviridae_AG099_hypothetical.p... 38.1 9e-06
Alpavirinae_Human_feces_D_015_Microviridae_AG018_hypothetical.p... 35.0 1e-04
Alpavirinae_Human_feces_C_043_Microviridae_AG0320_hypothetical.... 35.0 1e-04
Alpavirinae_Human_gut_22_017_Microviridae_AG0396_hypothetical.p... 28.5 0.017
> Alpavirinae_Human_feces_E_011_Microviridae_AG0387_hypothetical.protein.BACEGG.02723
Length=410
Score = 72.8 bits (177), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 46/119 (39%), Positives = 67/119 (56%), Gaps = 10/119 (8%)
Query 1 MAAQYYDAMASGHLKYQQAKSEITKRILMMAEAKGLQINNKAAEDTADGYIKALNAEYSA 60
MA Q + A ASG L QQ K+E+TK+++ MAE +G +I+NK A +TAD I AL +YSA
Sbjct 278 MANQSFQAFASGRLSLQQCKTEVTKQLMNMAETEGKKISNKIASETADQLIGALQWQYSA 337
Query 61 SYDINSPFKYGEDSYVPASVLKSRMDALNSKWQFDK-----RYWTEGLNALGTVGNAVG 114
+ + G Y + KSR +K Q D+ RYW G+ ++ +GN +G
Sbjct 338 ----DEMYSRGYAGYAREAG-KSRGKGDVAKGQLDEYDYSSRYWNTGIESINRIGNGIG 391
> Alpavirinae_Human_gut_33_017_Microviridae_AG0156_putative.VP2
Length=353
Score = 54.7 bits (130), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 44/119 (37%), Positives = 63/119 (53%), Gaps = 12/119 (10%)
Query 2 AAQYYDAMASGHLKYQQAKSEITKRILMMAEAKGLQINNKAAEDTADGYIKALNA--EYS 59
A+ YY+ M+ GHL Y QAK I IL A KG +++NK AE TAD I+A NA +
Sbjct 236 ASVYYNQMSQGHLNYNQAKKVIADEILTYARIKGQKLSNKVAEATADSLIRATNAANRSN 295
Query 60 ASYDINSPFKYGEDSYVPASV---LKSRMDALNSKWQFDKRYWTEGLNALGT-VGNAVG 114
A +D+ + KY + S+ +SR + K+ Y ++ L GT +GN VG
Sbjct 296 AEFDLEAA-KYNRERARSRSIEDWYRSRNEGKKYKY-----YDSDKLIHYGTSIGNTVG 348
> Alpavirinae_Human_feces_B_039_Microviridae_AG094_putative.VP2
Length=380
Score = 47.8 bits (112), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 41/141 (29%), Positives = 62/141 (44%), Gaps = 18/141 (13%)
Query 10 ASGHLKYQQAKSEITKRILMMAEAKGLQINNKAAEDTADGYIKALNAEYSASYDINSPFK 69
+ G L +Q ++EI + IL AEA G +I+N+ A +TAD IKA NA Y +S +
Sbjct 241 SQGALTEKQIQTEIQRAILASAEASGKKIDNRVASETADSLIKAANASNELQYR-DSTYD 299
Query 70 YGEDSYVPASVLKSRM--------DALNSKWQFDKRYWTEGLNALGTVGNAVGSVGNLRK 121
Y + K+ M A ++ Q YW +G++ G+ +
Sbjct 300 YKNVKLRKHTEYKTSMANQKAAEYGADLARKQGRTHYWESVARGIGSIAAGAGNFIGAFR 359
Query 122 PGA---------RNTYVYGNR 133
PGA RNT +Y R
Sbjct 360 PGANIFRNDYGPRNTTIYNGR 380
> Alpavirinae_Human_feces_A_034_Microviridae_AG0101_hypothetical.protein.BACPLE
Length=365
Score = 43.1 bits (100), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 39/128 (30%), Positives = 57/128 (45%), Gaps = 25/128 (20%)
Query 1 MAAQYYDAMASGHLKYQQAKSEITKRILMMAEAKGLQINNKAAEDTADGYIKALN----- 55
+ AQY++ + G +K +QAK+ + R+ + A G I+NK A TAD I A N
Sbjct 208 LGAQYWNMIRDGSIKEEQAKNLLATRLEIEARTAGQHISNKVARSTADSIIDATNTAKMN 267
Query 56 -AEYSASYDINSPFKYGEDSYVPASVLKSRMDALNSKWQFD--KRYWTEGLNALGT---- 108
A Y+ Y ++ D +MD +W D K W G+N G
Sbjct 268 EAAYNRGYS-----QFSND----VGYRTGKMD----RWSQDPVKARWDRGINNAGKFIDG 314
Query 109 VGNAVGSV 116
+ N VGSV
Sbjct 315 LSNIVGSV 322
> Alpavirinae_Human_gut_33_005_Microviridae_AG0182_hypothetical.protein.BACEGG.02723
Length=354
Score = 41.6 bits (96), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 36/121 (30%), Positives = 58/121 (48%), Gaps = 15/121 (12%)
Query 3 AQYYDAMASGHLKYQQAKSEITKRILMMAEAKGLQINNKAAEDTADGYIKALNAEYSASY 62
AQY++ + G + +QAK+ I R+ + A +G I+NK A+ TAD I A +
Sbjct 210 AQYWNMIREGVISEEQAKNLIASRLEIEARTQGQHISNKIAKSTADSIIDATR----TAK 265
Query 63 DINSPFKYGEDSYVPASVLKS-RMDALNSKWQFD--KRYWTEGLNALGT----VGNAVGS 115
+ + F G + ++ +MD +W D K W G+N G + NAVGS
Sbjct 266 ENEAAFNRGYSQFSNDVGFRTGKMD----RWLQDPVKARWDRGINNAGKFIDGLSNAVGS 321
Query 116 V 116
+
Sbjct 322 L 322
> Alpavirinae_Human_feces_D_031_Microviridae_AG0421_hypothetical.protein.BACEGG.02723
Length=418
Score = 40.0 bits (92), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 21/50 (42%), Positives = 32/50 (64%), Gaps = 0/50 (0%)
Query 11 SGHLKYQQAKSEITKRILMMAEAKGLQINNKAAEDTADGYIKALNAEYSA 60
+G L Q K++I ++ L+ A+A G ++NN+ AE AD KA+ AEY A
Sbjct 266 AGKLSEAQVKTQIKQQALLEAQAAGQKLNNRLAERLADYQFKAMAAEYRA 315
> Alpavirinae_Human_feces_D_008_Microviridae_AG099_hypothetical.protein.BACEGG.02723
Length=418
Score = 38.1 bits (87), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 20/50 (40%), Positives = 31/50 (62%), Gaps = 0/50 (0%)
Query 11 SGHLKYQQAKSEITKRILMMAEAKGLQINNKAAEDTADGYIKALNAEYSA 60
+G L Q K++I ++ L+ A+ G ++NN+ AE AD KA+ AEY A
Sbjct 266 AGKLSEAQVKTQIKQQALLEAQTVGQKLNNRLAERLADYQFKAMAAEYRA 315
> Alpavirinae_Human_feces_D_015_Microviridae_AG018_hypothetical.protein.BACEGG.02723
Length=427
Score = 35.0 bits (79), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 22/64 (34%), Positives = 35/64 (55%), Gaps = 2/64 (3%)
Query 2 AAQYYDAMASGHLKYQQAKSEITKRILMMAEAKGLQINNKAAEDTADGYIKALNAE--YS 59
AA Y + + G L +A+ +++ +L A A+GL I+N A +A G + A NA Y
Sbjct 263 AAHYEELINRGQLHVVEARELLSREVLNYARARGLNISNWVAAKSAKGLVYANNAANYYE 322
Query 60 ASYD 63
SY+
Sbjct 323 GSYN 326
> Alpavirinae_Human_feces_C_043_Microviridae_AG0320_hypothetical.protein.BACEGG.02723
Length=396
Score = 35.0 bits (79), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 20/55 (36%), Positives = 29/55 (53%), Gaps = 0/55 (0%)
Query 3 AQYYDAMASGHLKYQQAKSEITKRILMMAEAKGLQINNKAAEDTADGYIKALNAE 57
A+Y A L + ++EI I +AEA G +I+N+ A TA I A+N E
Sbjct 234 AEYSSITAGTELTKAKYRTEIANEIKTLAEANGQKISNEIARSTAQSLIDAMNKE 288
> Alpavirinae_Human_gut_22_017_Microviridae_AG0396_hypothetical.protein.BACPLE
Length=386
Score = 28.5 bits (62), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 38/118 (32%), Positives = 60/118 (51%), Gaps = 10/118 (8%)
Query 2 AAQYYDAMASGHLKYQQAKSEITKRILMMAEAKGLQINNKAAEDTADGYIKALNA--EYS 59
AA YY MA+G++ Y +AK + + L A +G I+N+ A A+ I A A E S
Sbjct 236 AADYYQRMAAGYVSYAEAKKALAEEALAAARTRGQNISNEVASRIAESQIAANIAANESS 295
Query 60 ASYDINSPFKYG--EDSYVPASV---LKSRMDALNSKWQFDKRYWTEGLNALG-TVGN 111
A+Y N + G +D+ ++ +SR + K+ FD W E ++G T+GN
Sbjct 296 AAYH-NEELRLGLPQDNARSKNIEEWYRSRNEKKRYKY-FDADKWVEYGTSIGNTIGN 351
Lambda K H a alpha
0.313 0.128 0.369 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 8809398