bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
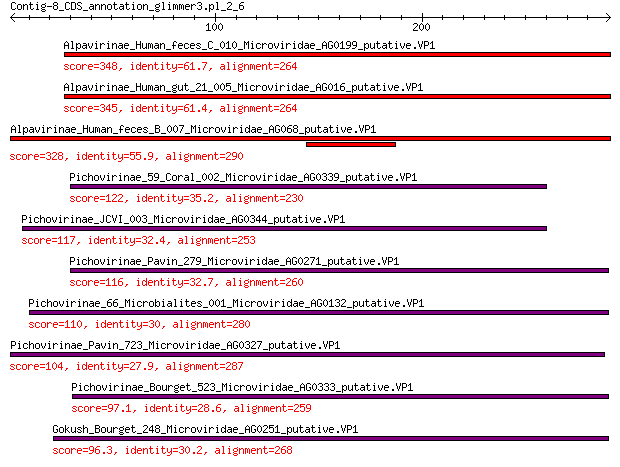
Query= Contig-8_CDS_annotation_glimmer3.pl_2_6
Length=290
Score E
Sequences producing significant alignments: (Bits) Value
Alpavirinae_Human_feces_C_010_Microviridae_AG0199_putative.VP1 348 7e-116
Alpavirinae_Human_gut_21_005_Microviridae_AG016_putative.VP1 345 9e-115
Alpavirinae_Human_feces_B_007_Microviridae_AG068_putative.VP1 328 1e-107
Pichovirinae_59_Coral_002_Microviridae_AG0339_putative.VP1 122 2e-33
Pichovirinae_JCVI_003_Microviridae_AG0344_putative.VP1 117 2e-31
Pichovirinae_Pavin_279_Microviridae_AG0271_putative.VP1 116 4e-31
Pichovirinae_66_Microbialites_001_Microviridae_AG0132_putative.VP1 110 6e-29
Pichovirinae_Pavin_723_Microviridae_AG0327_putative.VP1 104 7e-27
Pichovirinae_Bourget_523_Microviridae_AG0333_putative.VP1 97.1 2e-24
Gokush_Bourget_248_Microviridae_AG0251_putative.VP1 96.3 5e-24
> Alpavirinae_Human_feces_C_010_Microviridae_AG0199_putative.VP1
Length=731
Score = 348 bits (893), Expect = 7e-116, Method: Compositional matrix adjust.
Identities = 163/266 (61%), Positives = 204/266 (77%), Gaps = 8/266 (3%)
Query 27 SGISINDLRNVNAYQKFLELNMRKGYSYRDIIEGRFNVKVRYDELLMPEFFGGFSRDIEM 86
SG +I LR VNAYQKFLELNMRKG+SY+ I++GR+++ +R+DELLMPEF GG SR++ M
Sbjct 472 SGFTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSM 531
Query 87 HSISQTVDQDLDGSQ-TYAKALGSQSGIAGVRGDSGRALECFCDEESIVMGILIVTPLPV 145
++ QTVDQ S+ YA+ALGS++GIAGV G + +E FCDEES ++G+L VTP+P+
Sbjct 532 RTVEQTVDQQSGSSRGQYAEALGSKTGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPI 591
Query 146 YTQLLPKHFTYRGLLDHYQPEFNHIGFQPILYKEVCPLQAYS-DGPDTLSDVFGYNRPWY 204
YTQLLPK FTY GLLDHYQPEF+ IGFQPI YKE+CP+ + D + L+ FGY RPWY
Sbjct 592 YTQLLPKDFTYNGLLDHYQPEFDRIGFQPITYKEICPMNIVAGDSANQLNKTFGYQRPWY 651
Query 205 EYVQKYDQAHGLFRTNLSNFLMHRVFNQKPQLAQSFLVIDPAQVTDVFAVTKADDGTELA 264
EYV KYD AHGLFRTN+ NF+M RVF+ PQL Q FL++DP V VF+V TE
Sbjct 652 EYVAKYDSAHGLFRTNMKNFIMSRVFSGLPQLGQQFLLVDPDTVNQVFSV------TEYT 705
Query 265 DKIYGQIWFDCTAKLPISRVAIPRLD 290
DKI+G + F+ TA+LPISRVAIPRLD
Sbjct 706 DKIFGYVKFNATARLPISRVAIPRLD 731
> Alpavirinae_Human_gut_21_005_Microviridae_AG016_putative.VP1
Length=742
Score = 345 bits (886), Expect = 9e-115, Method: Compositional matrix adjust.
Identities = 162/265 (61%), Positives = 201/265 (76%), Gaps = 7/265 (3%)
Query 27 SGISINDLRNVNAYQKFLELNMRKGYSYRDIIEGRFNVKVRYDELLMPEFFGGFSRDIEM 86
SG +I LR VNAYQKFLELNMRKG+SY+ I++GR+++ +R+DELLMPEF GG SR++ M
Sbjct 484 SGFTIETLRYVNAYQKFLELNMRKGFSYKQIMQGRWDIDIRFDELLMPEFIGGISRELSM 543
Query 87 HSISQTVDQDLDGSQ-TYAKALGSQSGIAGVRGDSGRALECFCDEESIVMGILIVTPLPV 145
++ QTVDQ SQ YA+ALGS++GIAGV G + +E FCDEES ++G+L VTP+P+
Sbjct 544 RTVEQTVDQQSASSQGQYAEALGSKTGIAGVYGSTSNNIEVFCDEESYIIGLLTVTPVPI 603
Query 146 YTQLLPKHFTYRGLLDHYQPEFNHIGFQPILYKEVCPLQAYSDGPDTLSDVFGYNRPWYE 205
Y+QLLPK FTY GLLDHYQPEF+ IGFQPI YKEVCPL + ++ FGY RPWYE
Sbjct 604 YSQLLPKDFTYNGLLDHYQPEFDRIGFQPITYKEVCPLNFDISNMNDMNKTFGYQRPWYE 663
Query 206 YVQKYDQAHGLFRTNLSNFLMHRVFNQKPQLAQSFLVIDPAQVTDVFAVTKADDGTELAD 265
YV KYD AHGLFRT+L F+MHR F+ PQL Q FL++DP V VF+V TE D
Sbjct 664 YVAKYDNAHGLFRTDLKKFVMHRTFSGLPQLGQQFLLVDPDTVNQVFSV------TEYTD 717
Query 266 KIYGQIWFDCTAKLPISRVAIPRLD 290
KI+G + F+ TA+LPISRVAIPRLD
Sbjct 718 KIFGYVKFNATARLPISRVAIPRLD 742
> Alpavirinae_Human_feces_B_007_Microviridae_AG068_putative.VP1
Length=780
Score = 328 bits (841), Expect = 1e-107, Method: Compositional matrix adjust.
Identities = 162/290 (56%), Positives = 210/290 (72%), Gaps = 10/290 (3%)
Query 1 LKGVSYVELDNEVKLRQPRNLIDVVTSGISINDLRNVNAYQKFLELNMRKGYSYRDIIEG 60
LKGV+Y L + ++L+ VTSGISIND RNVNAYQ++LELN +G+SY++IIEG
Sbjct 501 LKGVNYTPLKAGEAINM-QSLVSPVTSGISINDFRNVNAYQRYLELNQFRGFSYKEIIEG 559
Query 61 RFNVKVRYDELLMPEFFGGFSRDIEMHSISQTVDQDLDGSQTYAKALGSQSGIAGVRGDS 120
RF+V VRYD L MPE+ GG +RDI ++ I+QTV+ GS Y +LGSQ+G+A G+S
Sbjct 560 RFDVNVRYDALNMPEYLGGITRDIVVNPITQTVETTDTGS--YVGSLGSQAGLATCFGNS 617
Query 121 GRALECFCDEESIVMGILIVTPLPVYTQLLPKHFTYRGLLDHYQPEFNHIGFQPILYKEV 180
++ FCDEESIVMGI+ V P+PVY +LPK TYR LD + PEF+HIG+QPI KE+
Sbjct 618 DGSISVFCDEESIVMGIMYVMPMPVYDSILPKWLTYRERLDSFNPEFDHIGYQPIYLKEL 677
Query 181 CPLQAYSDGPDTLSDVFGYNRPWYEYVQKYDQAHGLFRTNLSNFLMHRVFNQKPQLAQSF 240
+QA+ G D L+ VFGY RPWYEYVQK D+AHGLF ++L NF+M R F P+L +SF
Sbjct 678 AAIQAFESGKD-LNTVFGYQRPWYEYVQKVDRAHGLFLSSLRNFIMFRSFENAPELGKSF 736
Query 241 LVIDPAQVTDVFAVTKADDGTELADKIYGQIWFDCTAKLPISRVAIPRLD 290
V+ P V +VF+V TE++DKI GQI FDCTA+LPISRV +PRL+
Sbjct 737 TVMQPGSVNNVFSV------TEVSDKILGQIHFDCTAQLPISRVVVPRLE 780
Score = 25.0 bits (53), Expect = 0.91, Method: Compositional matrix adjust.
Identities = 13/43 (30%), Positives = 25/43 (58%), Gaps = 6/43 (14%)
Query 144 PVYTQLLPKHFTYRGLLDHYQPEFNHIGFQPILYKEVCPLQAY 186
PV+T+L+P + + R +PEF + F P+++ ++AY
Sbjct 37 PVFTELVPPNSSVR-----IKPEFG-LRFMPMMFPIQTKMKAY 73
> Pichovirinae_59_Coral_002_Microviridae_AG0339_putative.VP1
Length=522
Score = 122 bits (307), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 81/230 (35%), Positives = 110/230 (48%), Gaps = 11/230 (5%)
Query 30 SINDLRNVNAYQKFLELNMRKGYSYRDIIEGRFNVKVRYDELLMPEFFGGFSRDIEMHSI 89
SINDLR Q++LE N R G Y +II F V+ L PEF GG + I + +
Sbjct 280 SINDLRRAFRLQEWLERNARGGARYIEIIMAHFGVRSSDARLQRPEFLGGSATPITISEV 339
Query 90 SQTVDQDLDGSQTYAKALGSQSGIAGVRGDSGRALECFCDEESIVMGILIVTPLPVYTQL 149
QT + T G+ +G GV S + C+E ++GI+ V P Y Q
Sbjct 340 LQT-----SANNTEPTPQGNMAG-HGVSVGSSNYVSYKCEEHGYIIGIMSVMPKTAYQQG 393
Query 150 LPKHFTYRGLLDHYQPEFNHIGFQPILYKEVCPLQAYSDGPDTLSDVFGYNRPWYEYVQK 209
+PKHF D+Y P F +IG QPIL +E+ Y T ++ FGY + EY
Sbjct 394 IPKHFKKLDKFDYYWPSFANIGEQPILNEEL-----YHQNNATDTETFGYTPRYAEYKYI 448
Query 210 YDQAHGLFRTNLSNFLMHRVFNQKPQLAQSFLVIDPAQVTDVFAVTKADD 259
HG FR +L + M R+F KP L Q F+ + V VFAVT +
Sbjct 449 PSTVHGEFRDSLDFWHMGRIFGSKPTLNQDFIECNADDVERVFAVTAGQE 498
> Pichovirinae_JCVI_003_Microviridae_AG0344_putative.VP1
Length=505
Score = 117 bits (293), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 82/253 (32%), Positives = 123/253 (49%), Gaps = 15/253 (6%)
Query 7 VELDNEVKLRQPRNLIDVVTSGISINDLRNVNAYQKFLELNMRKGYSYRDIIEGRFNVKV 66
V DN+ P ++V ++ +I DLR Q++LEL R G Y + I+ F+VK
Sbjct 244 VGSDNDPFAYDPDGSLEVTST--TITDLRRAFKLQEWLELLARGGSRYIEQIKTFFDVKS 301
Query 67 RYDELLMPEFFGGFSRDIEMHSISQTVDQDLDGSQTYAKALGSQSGIAGVRGDSGRALEC 126
L PE+ G + + + I T +T G+ SG GV +G +
Sbjct 302 SDQRLQRPEYITGTKQPVIISEILNTT------GETAGLPQGNMSG-HGVAVGTGNVGKY 354
Query 127 FCDEESIVMGILIVTPLPVYTQLLPKHFTYRGLLDHYQPEFNHIGFQPILYKEVCPLQAY 186
+C+E ++GIL VTP Y Q +PKH+ D Y P+F HIG QPI+ E+ Y
Sbjct 355 YCEEHGFIIGILSVTPNTAYQQGIPKHYLRTDKYDFYWPQFAHIGEQPIVNNEI-----Y 409
Query 187 SDGPDTLSDVFGYNRPWYEYVQKYDQAHGLFRTNLSNFLMHRVFNQKPQLAQSFLVIDPA 246
+ P T + FGY + EY + G FR++L + R+F+ P L+Q F+ +D
Sbjct 410 AYDP-TGDETFGYTPRYAEYKYMPSRVAGEFRSSLDEWHAARIFSSLPTLSQDFIEVDAD 468
Query 247 QVTDVFAVTKADD 259
+FAVT DD
Sbjct 469 DFDRIFAVTDGDD 481
> Pichovirinae_Pavin_279_Microviridae_AG0271_putative.VP1
Length=503
Score = 116 bits (290), Expect = 4e-31, Method: Compositional matrix adjust.
Identities = 85/262 (32%), Positives = 125/262 (48%), Gaps = 23/262 (9%)
Query 30 SINDLRNVNAYQKFLELNMRKGYSYRDIIEGRFNVKVRYDELLMPEFFGGFSRDIEMHSI 89
+INDLR Q+FLE N R G Y + I+ F V+ L PE+ G + + I
Sbjct 263 TINDLRRAMRLQEFLEKNARGGTRYIENIKMHFGVQSSDSRLQRPEYITGTKAPVVVQEI 322
Query 90 SQTVD--QDLDGSQTYAKALGSQSGIAGVRGDSGRALECFCDEESIVMGILIVTPLPVYT 147
+ TV ++ G+ T KA QSG G +E ++GI+ + P P Y
Sbjct 323 TSTVKGIEEPQGNPT-GKATSLQSGHYG---------NYSVEEHGYIIGIMSIMPKPAYQ 372
Query 148 QLLPKHFTYRGLLDHYQPEFNHIGFQPILYKEVCPLQAYSDGPDTLSDVFGYNRPWYEYV 207
Q +PK + +LD Y P F +IG Q +L +E+ Y+ P+ VFGY + EY
Sbjct 373 QGIPKSYLKTDVLDFYWPSFANIGEQEVLQEEI---YGYTPTPNA---VFGYIPRYAEYK 426
Query 208 QKYDQAHGLFRTNLSNFLMHRVFNQKPQLAQSFLVIDPAQVTDVFAVTKADDGTELADKI 267
++ GLFRT L + + R+F P L+QSF+ + PA T +FA + D D I
Sbjct 427 YLQNRVCGLFRTTLDFWHLGRIFANIPTLSQSFIEVSPANQTRIFANQEDYD-----DNI 481
Query 268 YGQIWFDCTAKLPISRVAIPRL 289
Y + TA+ P+ P L
Sbjct 482 YIHVLNKMTARRPMPVFGTPML 503
> Pichovirinae_66_Microbialites_001_Microviridae_AG0132_putative.VP1
Length=518
Score = 110 bits (274), Expect = 6e-29, Method: Compositional matrix adjust.
Identities = 84/285 (29%), Positives = 136/285 (48%), Gaps = 28/285 (10%)
Query 10 DNEVKLRQPRN--LIDVVTSGISINDLRNVNAYQKFLELNMRKGYSYRDIIEGRFNVKVR 67
DN ++ +PR ++DV + IN +R Q++LE N R G Y + I F V+
Sbjct 257 DNPLQF-EPRGSLVVDVQADAVDINTVRRAFRLQEWLEKNARAGTRYIESILSHFGVRSS 315
Query 68 YDELLMPEFFGGFSRDI---EMHSISQTVDQDLDGSQTYAKALGSQSGIAGVRGDSGRAL 124
L PE+ GG ++ E+ + ++T D ++ A+G+ +G G+ G +
Sbjct 316 DARLQRPEYLGGTKGNMVISEVLATAETTDANV--------AVGTMAG-HGITAHGGNTI 366
Query 125 ECFCDEESIVMGILIVTPLPVYTQLLPKHFTYRGLLDHYQPEFNHIGFQPILYKEVCPLQ 184
+ C+E ++GI+ V P+ Y Q LP+ F+ LD+ P F +IG Q + KE L
Sbjct 367 KYRCEEHGWIIGIINVQPVTAYQQGLPREFSRFDRLDYPWPVFANIGEQEVYNKE---LY 423
Query 185 AYSDGPDTLSDVFGYNRPWYEYVQKYDQAHGLFRTNLSNFLMHRVFNQKPQLAQSFLVID 244
A S PD ++FGY + E K + G R L + + R+FN P L + F+ +
Sbjct 424 ANSPAPD---EIFGYIPRYAEMKFKNSRVAGEMRDTLDFWHLGRIFNNTPALNEEFIECN 480
Query 245 PAQVTDVFAVTKADDGTELADKIYGQIWFDCTAKLPISRVAIPRL 289
P T +FAVT + D I I+ + T + R IP +
Sbjct 481 PD--TRIFAVTDPAE-----DHIVAHIYNEVTVNRKLPRYGIPTI 518
> Pichovirinae_Pavin_723_Microviridae_AG0327_putative.VP1
Length=508
Score = 104 bits (259), Expect = 7e-27, Method: Compositional matrix adjust.
Identities = 80/287 (28%), Positives = 130/287 (45%), Gaps = 22/287 (8%)
Query 1 LKGVSYVELDNEVKLRQPRNLIDVVTSGISINDLRNVNAYQKFLELNMRKGYSYRDIIEG 60
+ G + V D + + ID+ + +INDLR Q++LE N R G Y + I
Sbjct 241 IGGSNAVNADGTPTMIAKTSTIDIEPT--TINDLRRAFKLQEWLEKNARGGTRYIENILT 298
Query 61 RFNVKVRYDELLMPEFFGGFSRDIEMHSISQTVDQDLDGSQTYAKALGSQSGIAGVRGDS 120
F V+ L PE+ G + + + T QD Q G+ +G G+ S
Sbjct 299 HFGVRSSDKRLQRPEYITGVKSPVVVSEVLNTTGQDGGLPQ------GNMAG-HGISVTS 351
Query 121 GRALECFCDEESIVMGILIVTPLPVYTQLLPKHFTYRGLLDHYQPEFNHIGFQPILYKEV 180
G++ +C+E ++GI+ V P Y Q +P+ F LD++ P F +IG Q + +E
Sbjct 352 GKSGSYYCEEHGYIIGIMSVMPKTAYQQGIPRTFLKTDSLDYFWPTFANIGEQEVAKQE- 410
Query 181 CPLQAYSDGPDTLSDVFGYNRPWYEYVQKYDQAHGLFRTNLSNFLMHRVFNQKPQLAQSF 240
L AY+ + D FGY + EY + G FRT+L+ + + R+F +P L F
Sbjct 411 --LYAYTANAN---DTFGYVPRYAEYKYMPSRVAGEFRTSLNYWHLGRIFATEPSLNSDF 465
Query 241 LVIDPAQVTDVFAVTKADDGTELADKIYGQIWFDCTAKLPISRVAIP 287
+ DP + +FAV + D +Y + A P+ + P
Sbjct 466 IECDPTK--RIFAVEDPE-----TDVLYCHVLNKIKAVRPMPKYGTP 505
> Pichovirinae_Bourget_523_Microviridae_AG0333_putative.VP1
Length=518
Score = 97.1 bits (240), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 74/259 (29%), Positives = 119/259 (46%), Gaps = 22/259 (8%)
Query 31 INDLRNVNAYQKFLELNMRKGYSYRDIIEGRFNVKVRYDELLMPEFFGGFSRDIEMHSIS 90
IN LR Q++LE N R G Y + I F VK L PE+ GG + + +
Sbjct 282 INSLRRAFRLQEWLERNARGGTRYIESILAHFGVKSSDARLQRPEYLGGSKGKMVISEVL 341
Query 91 QTVDQDLDGSQTYAKALGSQSGIAGVRGDSGRALECFCDEESIVMGILIVTPLPVYTQLL 150
T + L +G+ +G G+ G + +E ++GI+ VTP Y Q +
Sbjct 342 STAETTL--------PVGNMAG-HGISVSGGNEFKYNVEEHGWIIGIISVTPETAYQQGI 392
Query 151 PKHFTYRGLLDHYQPEFNHIGFQPILYKEVCPLQAYSDGPDTLSDVFGYNRPWYEYVQKY 210
+ + LD++ P F +IG Q +L KE+ Y++G + + + FGY + EY
Sbjct 393 HRSLSKLDRLDYFWPTFANIGEQEVLGKEI-----YANGIN-IGETFGYVPRYAEYKFLN 446
Query 211 DQAHGLFRTNLSNFLMHRVFNQKPQLAQSFLVIDPAQVTDVFAVTKADDGTELADKIYGQ 270
+ G RT+L + + R F+ P L +F+ DP+ T +FAV A+ D IYG
Sbjct 447 SRVAGEMRTSLDYWHLGRKFSAAPNLNGAFIECDPS--TRIFAVEDAE-----VDNIYGH 499
Query 271 IWFDCTAKLPISRVAIPRL 289
I+ + A + + P
Sbjct 500 IFNNIKAIRKMPKYGTPNF 518
> Gokush_Bourget_248_Microviridae_AG0251_putative.VP1
Length=546
Score = 96.3 bits (238), Expect = 5e-24, Method: Compositional matrix adjust.
Identities = 81/285 (28%), Positives = 119/285 (42%), Gaps = 31/285 (11%)
Query 22 IDVVTSGIS-------------INDLRNVNAYQKFLELNMRKGYSYRDIIEGRFNVKVRY 68
+ VVTSG+S IN LR QK LE + R G Y +II F V
Sbjct 272 LGVVTSGVSGLYADLSVATAATINQLRQSFQIQKLLERDARGGTRYTEIIRSHFGVISPD 331
Query 69 DELLMPEFFGGFSRDIEMHSISQTVDQDLDGSQTYAKALGSQSGIAGVRGDSGRALECFC 128
L PE+ GG S +I ++ I+QT + G+ T L + G
Sbjct 332 ARLQRPEYIGGGSTNININPIAQTSGTNASGTTTPMGTLAAM----GTALAHNHGFTYSS 387
Query 129 DEESIVMGILIVTPLPVYTQLLPKHFTYRGLLDHYQPEFNHIGFQPILYKEVCPLQAYSD 188
E +++G++ V Y Q L + ++ D Y P F +G Q +L KE+ Y
Sbjct 388 TEHGVIIGMVSVRADLTYQQGLQRMWSRSTRYDFYFPAFATLGEQAVLNKEI-----YVT 442
Query 189 GPDTLSDVFGYNRPWYEYVQKYDQAHGLFRTNLSNFL----MHRVFNQKPQLAQSFLVID 244
G +DVFGY W EY + LFR+ S + + + F P L +F + D
Sbjct 443 GNAGDNDVFGYQERWAEYRYYPSRISSLFRSTASGTIDAWHLAQKFTTTPTLNNTF-IQD 501
Query 245 PAQVTDVFAVTKADDGTELADKIYGQIWFDCTAKLPISRVAIPRL 289
V+ V AV A +G + + FDC P+ ++P L
Sbjct 502 TPPVSRVVAVGAAANGQQFIFDSF----FDCKKARPMPMYSVPGL 542
Lambda K H a alpha
0.322 0.141 0.422 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 24051702