bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-9_CDS_annotation_glimmer3.pl_2_3
Length=561
Score E
Sequences producing significant alignments: (Bits) Value
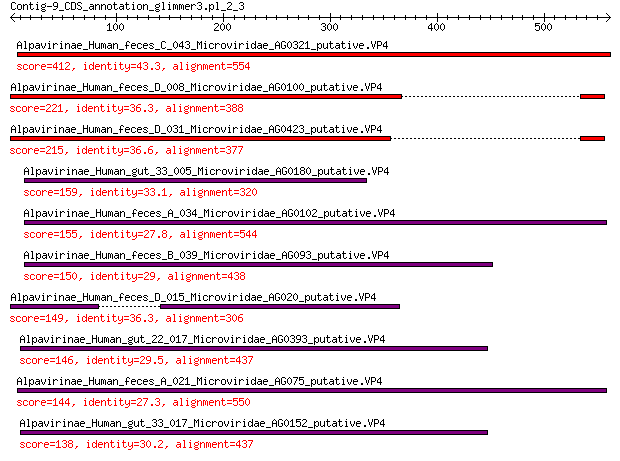
Alpavirinae_Human_feces_C_043_Microviridae_AG0321_putative.VP4 412 7e-139
Alpavirinae_Human_feces_D_008_Microviridae_AG0100_putative.VP4 221 7e-66
Alpavirinae_Human_feces_D_031_Microviridae_AG0423_putative.VP4 215 9e-64
Alpavirinae_Human_gut_33_005_Microviridae_AG0180_putative.VP4 159 5e-44
Alpavirinae_Human_feces_A_034_Microviridae_AG0102_putative.VP4 155 2e-42
Alpavirinae_Human_feces_B_039_Microviridae_AG093_putative.VP4 150 3e-41
Alpavirinae_Human_feces_D_015_Microviridae_AG020_putative.VP4 149 2e-40
Alpavirinae_Human_gut_22_017_Microviridae_AG0393_putative.VP4 146 1e-39
Alpavirinae_Human_feces_A_021_Microviridae_AG075_putative.VP4 144 6e-39
Alpavirinae_Human_gut_33_017_Microviridae_AG0152_putative.VP4 138 8e-37
> Alpavirinae_Human_feces_C_043_Microviridae_AG0321_putative.VP4
Length=546
Score = 412 bits (1060), Expect = 7e-139, Method: Compositional matrix adjust.
Identities = 240/563 (43%), Positives = 330/563 (59%), Gaps = 54/563 (10%)
Query 8 KYLAKRCYHGKNVINPYTGQVMYQPCGECPACLTRKASIRSMRVSLQKSLSKYTYFINPS 67
K L+K C HGKNV N YTGQ+MYQ CG+C ACL+R AS RS++V +Q SLSKY F+ +
Sbjct 9 KELSKTCLHGKNVYNKYTGQMMYQSCGKCEACLSRLASARSIKVGVQASLSKYVMFVTLT 68
Query 68 YDQKYVPKCQIFKVHPDDPDSTLYELVVKPRRRGDykmvqkvyskkykkvikkEVDAPLS 127
YD +VPKC+I+ ++ Y LVVKPR + + K+ LS
Sbjct 69 YDTYHVPKCKIYSNGDNN-----YVLVVKPRVKDYFYYETSDGQKRL---------KGLS 114
Query 128 YTDDFSYYFNASEEYIRDFREQATLNVNGKYPHLQDYYGYISRKDGQLFVKRLRKHIFKF 187
Y DDF F A ++YI +F++QA L+V G YPHL+D YGY+SRKD QLF+KR+RK I +
Sbjct 115 YDDDFRVEFKADKDYIENFKKQANLDVKGCYPHLKDMYGYLSRKDCQLFMKRVRKQIRNY 174
Query 188 AGKYEKVHIYLVSEYGPVHFRPHFHILLFTDSDRVAKNIGRIVNSSWKFGRCDWSASRGD 247
EK+H Y+V EY P HFRPHFHIL F +S+ ++++ G IV S WKFG DWS SRGD
Sbjct 175 TD--EKIHTYIVGEYSPKHFRPHFHILFFFNSNELSQSFGSIVRSCWKFGLVDWSQSRGD 232
Query 248 AESYVAGYVNSFSRLPYHLKQDDRVKPYSRFSNGFATSCFFDAKEAVRDSISRPIEETPL 307
AESYVAGYVNSF+RLPYHL ++ P+ RFSN F+ SCF DAK+ +RDS ++TP+
Sbjct 233 AESYVAGYVNSFARLPYHLGSSPKIAPFGRFSNHFSESCFDDAKQTLRDSFFG--QKTPV 290
Query 308 -SPFLNGVPSLINGKLLAIRPPRSVVDSCFLRYACNGRLSSHELYWLVRSVSTTLSRSFQ 366
+PFLNGV +L+NGKLL RP RS VDSCF R A +GRLSSHEL L+R+V + + +
Sbjct 291 FAPFLNGVTNLVNGKLLLTRPSRSCVDSCFFRKARDGRLSSHELLHLIRAVCHVVEQGKR 350
Query 367 TVRRDNPDATLMDVCRLHLRSIYSMSSRSVEDFLSIENQLYTCYYYAR--LESPTTDVRS 424
RD + T ++ R +++ S + E LS+ ++L T YY+R L D ++
Sbjct 351 FFARDGLNYTFLNHARFIVKACKSFRYQKREKLLSLPSRLVTLLYYSRVDLTKDIQDYKN 410
Query 425 DDLFDSDCMRLYRLFSCVGKFISFWDITPSSSYEYIYRTVDLSKWYYSRLSYHMLRSQYT 484
D+ ++ +YR+ +F+ FW I +E I R +D+ + YY L Y LR ++
Sbjct 411 PDVDEAMAQSIYRVLLYTMRFVRFWKIDKMPYHEGI-RLLDMCREYYRLLDYQYLRQRFQ 469
Query 485 dlvglvdldeelvdFLIAPVTEDT------PSSNGVSALEHHPPFESLVKSHPILSLAQA 538
+ ED P+ N L ++V SL +
Sbjct 470 ---------------FLEQCEEDLLDFYLHPTENFADYLP-----RTIVTGINQYSLERY 509
Query 539 KAKTDCDNRVKHREINERNLKYV 561
+ +KHREIN+ NL+YV
Sbjct 510 QKA------IKHREINDLNLQYV 526
> Alpavirinae_Human_feces_D_008_Microviridae_AG0100_putative.VP4
Length=532
Score = 221 bits (562), Expect = 7e-66, Method: Compositional matrix adjust.
Identities = 132/367 (36%), Positives = 202/367 (55%), Gaps = 18/367 (5%)
Query 1 MDKVGTIKYLAKRCYHGKNVINPYTGQVMYQPCGECPACLTRKASIRSMRVSLQKSLSKY 60
M++ I+ + C H NPYTG+++ PCG CPAC K+ + +V Q +S++
Sbjct 1 MNQQEFIQKVFSMCQHQVQTKNPYTGELIMVPCGTCPACRFNKSILSQNKVHAQSLISRH 60
Query 61 TYFINPSYDQKYVPKCQIFKVHPDDPDSTLYELVVKPRRRGDykmvqkvyskkykkvikk 120
YF+ +Y Q+Y+P ++ + D+ + R R K+K I+
Sbjct 61 VYFVTLTYAQRYIP---YYEYEIEALDADFLAITAHCRDRNPMYRTYTYRGTKHKLRIRG 117
Query 121 EVDAPLSYTDDFSYYFNASEEYIRDFREQATLNVNGKYPHLQDYYGYISRKDGQLFVKRL 180
+ S+ F+ + +Y + ++A L+ +GKYP L D Y+ D L++KR+
Sbjct 118 LASPKVK-----SFSFSVNRDYWTSYAQKANLSFDGKYPALSDRIPYLLHDDVSLYMKRV 172
Query 181 RKHIFKFAGKYEKVHIYLVSEYGPVHFRPHFHILLFTDSDRVAKNIGRIVNSSWKFGRCD 240
RK+I K G E +H Y+V EYGPV FRPHFH+LLF +SD +A++I RI S W+FGR D
Sbjct 173 RKYISKL-GINETIHTYVVGEYGPVTFRPHFHLLLFFNSDELAQSIVRIARSCWRFGRVD 231
Query 241 WSASRGDAESYVAGYVNSFSRLPYHLKQDDRVKPYSRFSNGFATSCFFDAKEAVRDSISR 300
SASRGDAE YV+ Y+NSFS +P H+++ ++P++RFSN F S F + +++ + S
Sbjct 232 CSASRGDAEDYVSSYLNSFSSIPLHIQEIRAIRPFARFSNKFGFSFF---ESSIKKAQSG 288
Query 301 PIEETPLSPFLNGVPSLINGKLLAIRPPRSVVDSCFLRYACNGRLSSHELYWLVRSVSTT 360
+E LNG NG I P R+++D+CF R A R HEL ++R V
Sbjct 289 DFDE-----ILNGKSLPYNGFNTTIFPWRAIIDTCFYRPALRRRSDIHELTEILRYVRNF 343
Query 361 LSR-SFQ 366
R +FQ
Sbjct 344 KQRPAFQ 350
Score = 23.1 bits (48), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 9/22 (41%), Positives = 14/22 (64%), Gaps = 0/22 (0%)
Query 534 SLAQAKAKTDCDNRVKHREINE 555
+L + K + R+KHREIN+
Sbjct 495 NLCRDKLHNEIRKRIKHREIND 516
> Alpavirinae_Human_feces_D_031_Microviridae_AG0423_putative.VP4
Length=532
Score = 215 bits (547), Expect = 9e-64, Method: Compositional matrix adjust.
Identities = 128/355 (36%), Positives = 196/355 (55%), Gaps = 17/355 (5%)
Query 1 MDKVGTIKYLAKRCYHGKNVINPYTGQVMYQPCGECPACLTRKASIRSMRVSLQKSLSKY 60
M++ I+ + C H NPYTG+++ PCG CPAC K+ + +V Q +S++
Sbjct 1 MNQQEFIQKVFSMCQHQVQTKNPYTGELIMVPCGTCPACRFNKSILSQNKVHAQSLVSRH 60
Query 61 TYFINPSYDQKYVPKCQIFKVHPDDPDSTLYELVVKPRRRGDykmvqkvyskkykkvikk 120
YF+ +Y Q+Y+P ++ + D+ + R K+K I+
Sbjct 61 VYFVTLTYAQRYIP---YYEYEIEALDADFLAITAHCCDRNPMYRTYTYRGTKHKLRIRG 117
Query 121 EVDAPLSYTDDFSYYFNASEEYIRDFREQATLNVNGKYPHLQDYYGYISRKDGQLFVKRL 180
L+ + S+ F+ + +Y + ++A L+ NGKYP L Y+ D L++KR+
Sbjct 118 -----LASPNVKSFSFSVNRDYWTSYAQKANLSFNGKYPALSGRIPYLLHGDVSLYMKRV 172
Query 181 RKHIFKFAGKYEKVHIYLVSEYGPVHFRPHFHILLFTDSDRVAKNIGRIVNSSWKFGRCD 240
RK+I K G E +H Y+V EYGP FRPHFH+LLF DSD +A+NI RI +S W+FGR D
Sbjct 173 RKYISKL-GINETIHTYIVGEYGPSSFRPHFHLLLFFDSDELAQNIIRIASSCWRFGRVD 231
Query 241 WSASRGDAESYVAGYVNSFSRLPYHLKQDDRVKPYSRFSNGFATSCFFDAKEAVRDSISR 300
SASRGDAE YV+ Y+NSFS +P H+++ ++P++RFSN F S F + +++ + S
Sbjct 232 CSASRGDAEDYVSSYLNSFSSIPLHIQEIRAIRPFARFSNKFGYSFF---ESSIKKAQSG 288
Query 301 PIEETPLSPFLNGVPSLINGKLLAIRPPRSVVDSCFLRYACNGRLSSHELYWLVR 355
+E LNG NG I P R+++D+CF R A HEL ++R
Sbjct 289 NFDEI-----LNGKSLPYNGFDTTIFPWRAIIDTCFYRPALRRHSDIHELTEILR 338
Score = 25.0 bits (53), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 10/22 (45%), Positives = 15/22 (68%), Gaps = 0/22 (0%)
Query 534 SLAQAKAKTDCDNRVKHREINE 555
SL + K ++ R+KHREIN+
Sbjct 495 SLCRDKLHSEIRKRIKHREIND 516
> Alpavirinae_Human_gut_33_005_Microviridae_AG0180_putative.VP4
Length=536
Score = 159 bits (402), Expect = 5e-44, Method: Compositional matrix adjust.
Identities = 106/330 (32%), Positives = 168/330 (51%), Gaps = 38/330 (12%)
Query 14 CYHGKNVINPYTGQVMYQPCGECPACLTRKASIRSMRVSLQKSLSKYTYFINPSYDQKYV 73
C H + N YTG +Y CG C CL+ KA + +R ++Q + S+ +F+ +Y +++
Sbjct 12 CLHPVIIKNKYTGDPIYVECGTCEVCLSNKAIQKELRCNIQLASSRCCFFVTLTYATEHI 71
Query 74 PKCQIFKVHPDDPDSTLYELVVKPRRRGDykmvqkvyskkykkvikkEVDAPLSYTDD-F 132
P + +K++ DS Y L PR Y V + + +S+ D+ F
Sbjct 72 PVARFYKLN----DS--YHLCCVPRDH------------VYTYVTSQGYNRKMSFCDEEF 113
Query 133 SYYFNASEEYIRDFREQATLNVNG--------KYPHLQDYYGYISRKDGQLFVKRLRKHI 184
Y N +E + ++ L+ KYP++ D Y++ +D QLF KR+ + I
Sbjct 114 DYPTNLRDEAVTALLDKTHLDRTVYPDGRSVVKYPNMGDLLPYLNYRDVQLFHKRINQQI 173
Query 185 FKFAGKYEKVHIYLVSEYGPVHFRPHFHILLFTDSDRVAKNIGRIVNSSWKFGRCDWSAS 244
K+ EK++ Y V EYGP FRPHFH+L F DS+R+A+ G++V+ +W+FG D
Sbjct 174 KKYTD--EKIYSYTVGEYGPKTFRPHFHLLFFFDSERLAQVFGQLVDKAWRFGNSDTQRV 231
Query 245 RGDAESYVAGYVNSFSRLPYHLKQDDRVKPYSRFSNGFATSCFFDAKEAVRDSISRPIE- 303
A SYVAGY+NS LP + + ++ P+ RFS FA F +A +P E
Sbjct 232 WSSASSYVAGYLNSSHCLPEFYRCNRKIAPFGRFSQYFAERPFIEA--------FKPEEN 283
Query 304 ETPLSPFLNGVPSLINGKLLAIRPPRSVVD 333
E F+NG+ + GK RP RS+++
Sbjct 284 EKVFDKFVNGIYLSLGGKPTLCRPKRSLIN 313
> Alpavirinae_Human_feces_A_034_Microviridae_AG0102_putative.VP4
Length=539
Score = 155 bits (391), Expect = 2e-42, Method: Compositional matrix adjust.
Identities = 151/564 (27%), Positives = 246/564 (44%), Gaps = 68/564 (12%)
Query 14 CYHGKNVINPYTGQVMYQPCGECPACLTRKASIRSMRVSLQKSLSKYTYFINPSYDQKYV 73
C H + + N YTG +Y PCG C C+ KA ++ ++Q + S+Y FI +Y +Y+
Sbjct 12 CLHPRVIKNKYTGDPIYVPCGTCEFCVHNKAIKAELKCNVQLASSRYCEFITLTYSTEYL 71
Query 74 PKCQIFKVHPDDPDSTLYELVVKPRRRGDykmvqkvyskkykkvikkEVDAPLSYTDD-F 132
P + ++ P RG+ + Y + +S+ D F
Sbjct 72 PVGKFYQ----GP-------------RGEVRFCSLPRDFVYSYKTVQGYTRNISFNDKTF 114
Query 133 SYYFNASEEYIRDFREQATLNVNG--------KYPHLQDYYGYISRKDGQLFVKRLRKHI 184
+ S + +++A L+ P+++ GY++ D QLF KRL ++I
Sbjct 115 DFDTQLSWSSAQLLQKKAHLHYTSFPDGRCIYNRPYMEGLVGYLNYHDIQLFFKRLNQNI 174
Query 185 FKFAGKYEKVHIYLVSEYGPVHFRPHFHILLFTDSDRVAKNIGRIVNSSWKFGRCDWSAS 244
+ EK++ Y+V EYGP FRPHFHILLF DS ++ ++I + V+ SW+FG D
Sbjct 175 RRITN--EKIYYYVVGEYGPTTFRPHFHILLFHDSRKLRESIRQFVSKSWRFGDSDTQPV 232
Query 245 RGDAESYVAGYVNSFSRLPYHLKQDDRVKPYSRFSNGFATSCFFDAKEAVRDSISRPIE- 303
A YVAGYVNS + LP K +KP+ RFS FA S F + + +P E
Sbjct 233 WSSASCYVAGYVNSTACLPDFFKNSRHIKPFGRFSVNFAESAF--------NEVFKPEEN 284
Query 304 ETPLSPFLNGVPSLINGKLLAIRPPRSVVDSCFLRYACNGRLSSHELYWLVRSVSTTLSR 363
E S F +G +NGK +RP RS ++ + R S H +++ +SR
Sbjct 285 EEIFSLFYDGRVLNLNGKPTIVRPKRSHINRLYPRLD----KSKHATVVDDIRIASVVSR 340
Query 364 SFQTVRRDNPDATLMDVCRLHL-RSIYSMSSRSVEDFLSIE---NQLYTCYYYARLESPT 419
Q V + + +V L + +Y + R +E S++ ++L Y RL +
Sbjct 341 LPQVVAK---FGFIDEVSDFELSKRVYYLIRRHLEVDCSLDYASDELRVIYNACRL---S 394
Query 420 TDVRSDDLFDSDCMRLYRLFSCVGKFISFWDITPSSSYEY---IYRTVDLSKWYY---SR 473
+ D +S C +YRL + W P S + ++R V +Y SR
Sbjct 395 LYINFSD--ESGCAAIYRLLLQYRNLVKNWITAPVGSVAFTGQLHRAVRAIHSFYDYCSR 452
Query 474 LSYHMLRSQYTdlvglvdldeelvdFLIAPVTEDTPSSNGVSALEHHPPFESLVKSHPIL 533
S H + + L + P+T+ V + F +V +L
Sbjct 453 RSLHDQLVKVEKWSNDSYVRANLSIYYFYPLTD-------VDIM--MKSFSEIVFDSQVL 503
Query 534 SLAQAKAKTDCDNRVKHREINERN 557
+ A D R+KH+ +N++N
Sbjct 504 RASYADYAADNRERIKHKALNDKN 527
> Alpavirinae_Human_feces_B_039_Microviridae_AG093_putative.VP4
Length=559
Score = 150 bits (380), Expect = 3e-41, Method: Compositional matrix adjust.
Identities = 127/461 (28%), Positives = 207/461 (45%), Gaps = 43/461 (9%)
Query 14 CYHGKNVINPYTGQVMYQPCGECPACLTRKASIRSMRVSLQKSLSKYTYFINPSYDQKYV 73
C H + N Y G + CG+C C+ ++A SMRV S KY+YF+ +YD +++
Sbjct 14 CQHRSFITNRYNGARITVDCGQCDYCIHKRAQKASMRVKTAGSAFKYSYFVTLTYDNEHI 73
Query 74 P--KCQIFKVHPDD----------PDSTLYELVVKPRRRGDykmvqkvyskkykkvikkE 121
P C++ +D D + V R D M++ ++ + +
Sbjct 74 PLMNCKVLHSEYEDVVGISGDIHFGDEYHKYIPVSEYRCDDNSMLRHIFFE--------Q 125
Query 122 VDAPLSYTDDFSYYFNASEEY------IRDFREQATLNVNGKYPHLQDY-----YGYISR 170
V + + + Y + + IR F + YP + Y +++
Sbjct 126 VQGTVPFDREIKEYVPVKDNWFLSMAAIRSFIYKTQSVDKTDYPASEQYGRDNLIPFLNY 185
Query 171 KDGQLFVKRLRKHIFKFAGKYEKVHIYLVSEYGPVHFRPHFHILLFTDSDRVAKNIGRIV 230
D Q ++KRLRK++F+ GKYE H Y V EYGPVHFRPH+H+LLFT+SD+V++ +
Sbjct 186 VDVQNYIKRLRKYLFQQLGKYETFHFYAVGEYGPVHFRPHYHLLLFTNSDKVSEVLRYCH 245
Query 231 NSSWKFGRCDWSASRGDAESYVAGYVNSFSRLPYHLKQDDRVKPYSRFSNGFATSCFFDA 290
+ SWK GR D+ S G A SYVA YVNS P + +P SR S GF F
Sbjct 246 DKSWKLGRSDFQRSAGGAGSYVASYVNSLCSAPLLYRSCRAFRPKSRASVGF----FEKG 301
Query 291 KEAVRDSISRPIEETPLSPFLNGVPSLINGKLLAIRPPRSVVDSCFLRYACNGRLSSHEL 350
+ V D E + +NG NG + PP S + + R++ +
Sbjct 302 CDFVEDDDPYAQIEQKIDSVVNGRVYNFNGVSVRSTPPLSYIRTLLPRFSSARNDDGIAI 361
Query 351 YWLVRSVSTTLSRSFQTVRRDNPDATLMDVCRLHLRSIYSMSSRSVEDFLSIENQLYTCY 410
++ +V +T R + D +++ + R + + + V L+ +++L
Sbjct 362 ARILYAVHSTPKRIARFGFADYKQDSILSLVRTYYQYL------KVNPILTDDDKL--IL 413
Query 411 YYARLESPTTDVRSDDLFDSDCMRLYRLFSCVGKFISFWDI 451
+ +R + + SD +S +LYRLF V KF W +
Sbjct 414 HASRCLTRFVNCSSDVDIESYINKLYRLFLYVYKFFRNWHL 454
> Alpavirinae_Human_feces_D_015_Microviridae_AG020_putative.VP4
Length=563
Score = 149 bits (375), Expect = 2e-40, Method: Compositional matrix adjust.
Identities = 82/227 (36%), Positives = 125/227 (55%), Gaps = 12/227 (5%)
Query 141 EYIRDFREQATLNVNGKYPHLQDYYGYISRKDGQLFVKRLRKHIFKFAGKYEKVHIYLVS 200
E +++ + NG +P + Y++ +D QLF KRLRK++ K GKYEK+H Y+VS
Sbjct 183 EIAESLKKKNNTDANGAFPQFKGLLKYVNIRDYQLFAKRLRKYLSKKVGKYEKIHSYVVS 242
Query 201 EYGPVHFRPHFHILLFTDSDRVAKNIGRIVNSSWKFGRCDWSASRGDAESYVAGYVNSFS 260
EY P FRPHFHIL F DSD +AKN + V SW+ GR D +R A SYV+ Y+NS
Sbjct 243 EYSPKTFRPHFHILFFFDSDEIAKNFRQAVYQSWRLGRVDTQLAREQANSYVSNYLNSVV 302
Query 261 RLPYHLKQDDRVKPYSRFSNGFATSCFFDAKEAVRDSISRPIEETPLSPFLNGVPSLING 320
+P+ K ++P SRFSN F F + KE +R + + + +G+ + N
Sbjct 303 SIPFVYKAKKSIRPRSRFSNLFG---FEEVKEGIRKA------QDKRAALFDGLSYISNQ 353
Query 321 KLLAIRPPRSVVDSCFLRYA-CNGRL--SSHELYWLVRSVSTTLSRS 364
K + P S++D F R+ +G S ++Y +V+ + +R+
Sbjct 354 KFVRYVPSGSLIDRLFPRFTYYDGSFLRRSSQIYGVVQQILRLFARN 400
Score = 57.0 bits (136), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 29/82 (35%), Positives = 44/82 (54%), Gaps = 2/82 (2%)
Query 1 MDKVGTIKYLAKRCYHGKNVINPYTGQVMYQPCGECPACLTRKASIRSMRVSLQKSLSKY 60
MDK KY RC H + + N YTG + CG+CP CL +KA + + K +Y
Sbjct 1 MDK-KVFKYF-NRCEHPQVIQNKYTGDYVKVDCGQCPYCLIKKADRSTQKCDFVKYNHRY 58
Query 61 TYFINPSYDQKYVPKCQIFKVH 82
YF+ +Y+ +YVPK + ++
Sbjct 59 CYFVTLTYNTQYVPKMSLTQIE 80
> Alpavirinae_Human_gut_22_017_Microviridae_AG0393_putative.VP4
Length=562
Score = 146 bits (369), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 129/475 (27%), Positives = 207/475 (44%), Gaps = 80/475 (17%)
Query 10 LAKRCYHGKNVINPYTGQVMYQPCGECPACLTRKASIRSMRVSLQKSLSKYTYFINPSYD 69
L RC H + V+N YT + + CG CP+C+ R++S+++ ++ + +Y YF+ +Y
Sbjct 10 LVTRCQHPRTVVNKYTHEPVVVSCGHCPSCILRRSSVQTNLLTTYSAQFRYVYFVTLTYA 69
Query 70 QKYVPKCQIFKVH-------------------PDDPDSTLYELVVKPRRRGDykmvqkvy 110
++P ++ V DP++ L+ PR
Sbjct 70 PSFLPTLEVSVVETCTDDIADVSCVPNIDELDASDPNTYLFGFRSVPR------SSSVKL 123
Query 111 skkykkvikkEVDAPLSYTDDFSYYFNASEEYIRDFREQATLNVNGKYPH-LQDYYGYIS 169
+ K+ D SY + + L++ GK H + + YI
Sbjct 124 KSSTVERTFKDPDVKFSYP----------------MKPKELLSILGKINHNVPNRIPYIC 167
Query 170 RKDGQLFVKRLRKHIFKFAGKYEKVHIYLVSEYGPVHFRPHFHILLFTDSDRVAKNIGRI 229
+D LF+KRLR + EK+ Y VSEYGP FRPH+H+LLF++S+R ++ +
Sbjct 168 NRDLDLFLKRLRSYYLD-----EKLRYYAVSEYGPTSFRPHWHLLLFSNSERFSRTVCEN 222
Query 230 VNSSWKFGRCDWSASRGDAESYVAGYVNSFSRLP-YHLKQDDRVKPYSRFSNGFATSCFF 288
V+ +W +GRCD S SRG A YVA YVNSF LP ++ + V+P S S GF S F
Sbjct 223 VSKAWSYGRCDASLSRGFAAPYVASYVNSFVALPDFYTQMPKVVRPKSFHSIGFTESNLF 282
Query 289 DAKEAVRDSISRPIEETPLSPFLNGVPSLINGKLLAIRPPRSVVDSCFLRYACNGRLSSH 348
K V + ++E LNGV +G I+P + F R++ R S
Sbjct 283 PRKVRVAE-----VDEI-TDKCLNGVRVERDGYFRTIKPTWPYLLRLFPRFSDAIRKSPS 336
Query 349 ELYWLVRSVSTTLSRSFQTVRRD--------NPDATLMDVCRLHLRSIYSMS-SRSVEDF 399
+Y L+ + T R ++ D + +++ C+ +L + + S +F
Sbjct 337 SIYQLLFAAFTAPERVIRSGCADIGCDPFGESSKQSILSFCKQYLNYVDNYGKSNEHRNF 396
Query 400 LSIENQ--------LYTCYYYARLESPTTDVRSDDLFDSDCMRLYRLFSCVGKFI 446
LS + L C Y ++ TT S R+YR F + KFI
Sbjct 397 LSPQASLPHSDVLILSECRLYDGVDLETTHRVS---------RVYRFFLGISKFI 442
> Alpavirinae_Human_feces_A_021_Microviridae_AG075_putative.VP4
Length=547
Score = 144 bits (363), Expect = 6e-39, Method: Compositional matrix adjust.
Identities = 150/578 (26%), Positives = 245/578 (42%), Gaps = 68/578 (12%)
Query 8 KYLAKR------CYHGKNVINPYTGQVMYQPCGECPACLTRKASIRSMRVSLQKSLSKYT 61
+YL+ R C + + ++ V+ C EC CL +K+ ++ V++ KY
Sbjct 4 QYLSSRFAPMSDCNQPRYLRQLHSDDVLSIGCNECLPCLLKKSRNDTLDVTISTQTFKYI 63
Query 62 YFINPSYDQKYVPKCQIFKVHPDDPDSTLY--ELVVKP-------RRRGDykmvqkvysk 112
+F+N Y ++P + H P+ + E +V+P +RR D +V +S
Sbjct 64 FFVNLDYATPFIPIMR----HEYIPERKVIYCESMVRPSVKIPTNKRRKDGSVVFTTFSF 119
Query 113 kykkvikkEVDAPLSYTDDFSYYFNASEEYIRDFREQATLNVNGKYPHLQDYYGYISRKD 172
+ + P +T SEE R F + L NGKYP +D + Y+SR D
Sbjct 120 RAA-----DETRPFCFTAPCE-----SEEEYRLFVRKVNLTANGKYPMYKDCFPYLSRYD 169
Query 173 GQLFVKRLRKHIFKFAGKYEKVHIYLVSEYGPVHFRPHFHILLFTDSDRVAKNIGRIVNS 232
LF+KRLR K G+ +H Y+V EYGPVHFRPH+HI++ ++ R A+++ V+
Sbjct 170 VALFMKRLRNLFLKKYGQSIYMHSYIVGEYGPVHFRPHYHIIISSNDPRFAESLEHFVDK 229
Query 233 SWKFGRCDWSASRGDAESYVAGYVNSFSRLPYHLKQDDRVKPYSRFSNGFATSCFFDAKE 292
+W FG G+ SYVA YVNS LP ++ V+P+SR GF F
Sbjct 230 AWFFGGASAEIPDGNCASYVASYVNSSVSLPVLFRKHRLVRPFSRKCRGFLMERFAPDSL 289
Query 293 AVRDSISRPIEETPLSPFLNGVPSLINGKLLAIRPPRSVVDSCFLRYACNGRLSSHELYW 352
+ R I NG I + P RS D + R GR +L
Sbjct 290 PFGERTDRDI---------NGTLCSIGDASFQLYPRRSYRDRVYPRLFKVGRFDLDQLL- 339
Query 353 LVRSVSTTLSRSFQTVRRDNPDATLMDVCRLHLRSIYSMSSRSVEDFLSIENQLYTCYYY 412
+VS ++R F+T+ + T ++ R LR + + E L Y+
Sbjct 340 ---TVSRIITRIFRTICKQGRQRTAAELARSILRYVQFGRDPNTE-------VLRQFIYF 389
Query 413 ARLESPTTDVRSDD--LFDSDCMRLYRLFSCVGKFISFWDITPSSSYEYIYRTVDLS--- 467
+ P R+ LF+ R YR+ + FW + + R ++L+
Sbjct 390 NETDIPLARSRATRQMLFN----RSYRIAYNYCSLVRFWTSCDICGFGFD-REIELAIRN 444
Query 468 -KWYYSRLSYHMLRSQYTdlvglvdldeelvdFLIAPVTEDTPSSNGVSALEHHP----- 521
YYS Y L +QY D++ +D + + + D V + +H
Sbjct 445 VNRYYSETEYRYLVNQYEDILRFIDSEFYRGEVDDLLMYYDRSPGRFVVSKQHRKVSMSD 504
Query 522 --PFESLVKSHPILSLAQAKAKTDCDNRVKHREINERN 557
P S +K++ ++ + N +KH+E+N+ N
Sbjct 505 TFPV-STIKTNRLVQAVDEECHCRIQNSIKHKELNDLN 541
> Alpavirinae_Human_gut_33_017_Microviridae_AG0152_putative.VP4
Length=565
Score = 138 bits (347), Expect = 8e-37, Method: Compositional matrix adjust.
Identities = 132/476 (28%), Positives = 209/476 (44%), Gaps = 82/476 (17%)
Query 10 LAKRCYHGKNVINPYTGQVMYQPCGECPACLTRKASIRSMRVSLQKSLSKYTYFINPSYD 69
L RC + + V+N YT + + CG CP+C+ R++ I++ ++ + +Y YF+ +Y
Sbjct 10 LVTRCQNPRTVVNKYTHEPVVVSCGACPSCVLRRSGIQTNLLTTYSAQFRYVYFVTLTYA 69
Query 70 QKYVPKCQIFKVH-------------------PDDPDSTLYELVVKPRRRGDykmvqkvy 110
++P ++ + P D + L+ PR V+
Sbjct 70 PCFLPTLEVSVIETCTDDIADVSSVPDINDLDPCDNNRYLFGFCSVPR----SASVKLKN 125
Query 111 skkykkvikkEVDAPLSYTDDFSYYFNASEEYIRDFREQATLNVNGKYPHLQDYYGYISR 170
S + EV FSY E + ++ NV + P YI
Sbjct 126 STVERTFKDPEVR--------FSYPMKPKE--LLSILDKINHNVPNRIP-------YICN 168
Query 171 KDGQLFVKRLRKHIFKFAGKYEKVHIYLVSEYGPVHFRPHFHILLFTDSDRVAKNIGRIV 230
+D LF+KRLR + YEK+ Y VSEYGP +RPH+H+LLF++S++ +K I V
Sbjct 169 RDLDLFLKRLRSYY-----PYEKLRYYAVSEYGPTSYRPHWHLLLFSNSEQFSKTILENV 223
Query 231 NSSWKFGRCDWSASRGDAESYVAGYVNSFSRLP-YHLKQDDRVKPYSRFSNGFATSCFFD 289
+ +W +GRCD S SRG A YVA YVNSF LP ++ + ++P S S GF S F
Sbjct 224 SKAWSYGRCDASLSRGFAAPYVASYVNSFVALPSFYTEMPRFLRPKSFHSIGFTESNLFP 283
Query 290 AKEAVRDSISRPIEETPLSPFLNGVPSLINGKLLAIRPPRSVVDSCFLRYACNGRLSSHE 349
K + + I+E LNGV +G ++P + F R++ R S
Sbjct 284 RKVRISE-----IDEV-TDKCLNGVRVERDGYFRILKPSWPYLLRLFPRFSDAIRKSPSS 337
Query 350 LYWLVRSVSTTLSRSFQTVRRD--------NPDATLMDVCRLHLRSIYSMS-SRSVEDFL 400
+Y L+ + T +R ++ D N +L+ C+ +L + + S ++FL
Sbjct 338 IYQLLFAAFTAPARVIRSGCADIGCDPFVENSKQSLLSFCKQYLNYVDNHGKSNEFKNFL 397
Query 401 SIENQLYTCYYYARLESPTTDV------RSDDLFDSDCM----RLYRLFSCVGKFI 446
S L P +DV R D D + R+YR F + KFI
Sbjct 398 SPHADL-----------PHSDVLILTECRLYDGVDLEAAHRLSRVYRFFLGIAKFI 442
Lambda K H a alpha
0.322 0.137 0.422 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 51467408