bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-12_CDS_annotation_glimmer3.pl_2_1
Length=354
Score E
Sequences producing significant alignments: (Bits) Value
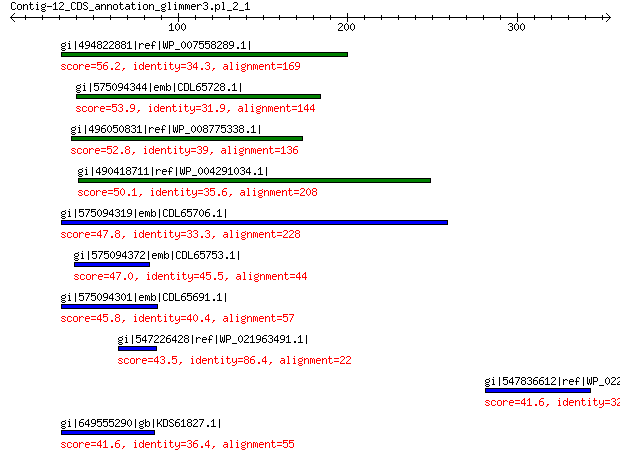
gi|494822881|ref|WP_007558289.1| hypothetical protein 56.2 1e-05
gi|575094344|emb|CDL65728.1| unnamed protein product 53.9 7e-05
gi|496050831|ref|WP_008775338.1| predicted protein 52.8 2e-04
gi|490418711|ref|WP_004291034.1| hypothetical protein 50.1 0.001
gi|575094319|emb|CDL65706.1| unnamed protein product 47.8 0.008
gi|575094372|emb|CDL65753.1| unnamed protein product 47.0 0.012
gi|575094301|emb|CDL65691.1| unnamed protein product 45.8 0.033
gi|547226428|ref|WP_021963491.1| putative uncharacterized protein 43.5 0.16
gi|547836612|ref|WP_022244433.1| putative uncharacterized protein 41.6 0.45
gi|649555290|gb|KDS61827.1| hypothetical protein M095_3809 41.6 0.61
>gi|494822881|ref|WP_007558289.1| hypothetical protein [Bacteroides plebeius]
gi|198272097|gb|EDY96366.1| hypothetical protein BACPLE_00802 [Bacteroides plebeius DSM 17135]
Length=344
Score = 56.2 bits (134), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 58/178 (33%), Positives = 87/178 (49%), Gaps = 37/178 (21%)
Query 31 KHQLEMQRIQNEWASSESQKSRDFAKSMFDATNEWNSAKNQRARLEEAGLNPYLMMNggs 90
+ QLE Q I+ EW M++A NE+NSA +QR RLEEAGLNPY+MM+GGS
Sbjct 59 REQLERQ-IEQEW-------------DMWNAENEYNSASSQRKRLEEAGLNPYMMMDGGS 104
Query 91 agtasstsantvsgasgsggsPYQFTPTNMIG-----DVAS-YAAAMKSMSDARK---TN 141
AG+ASS ++ A P +M G +AS + A +K+ D R N
Sbjct 105 AGSASSMTSPAAQPAVVPQMQGATMQPADMSGLSGLRGIASEFIATLKAQEDIRGQQLIN 164
Query 142 TESDLLDQYGAATYESRIKKTMADTYFTQRQADVAIAQRANLLLRNDAQEVLNMYLPE 199
++ +QY A + ++KT ++ F + Q Q+++N + PE
Sbjct 165 EGQEIENQYKADKLLADLEKTRTESGFVRSQT--------------KGQDIMNRFRPE 208
>gi|575094344|emb|CDL65728.1| unnamed protein product [uncultured bacterium]
Length=368
Score = 53.9 bits (128), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 46/146 (32%), Positives = 68/146 (47%), Gaps = 13/146 (9%)
Query 40 QNEWASSESQKSRDFAKSMFDATNEWNSAKNQRARLEEAGLNPYLMM-Nggsagtassts 98
Q EW S E+QK RDF M++ NE+N Q RLEEAG+NP+ M N A SS S
Sbjct 49 QMEWQSQEAQKQRDFQLDMWNRNNEYNKPDEQMKRLEEAGINPWQSMGNSSVASGNSSLS 108
Query 99 antvsgasgsggsPYQFTPTNMIGDV-ASYAAAMKSMSDARKTNTESDLLDQYGAATYES 157
+ S + + +P + DV A A K+ +D + NT ++
Sbjct 109 QPSGFVPSPAHAASSSLSPLGLAADVLGKIAQANKAGADTNRVNT-----------LLQT 157
Query 158 RIKKTMADTYFTQRQADVAIAQRANL 183
++K +A+ F QA + +NL
Sbjct 158 ELEKLIAEKNFVSLQAARQAIENSNL 183
>gi|496050831|ref|WP_008775338.1| predicted protein [Bacteroides sp. 2_2_4]
gi|229448895|gb|EEO54686.1| hypothetical protein BSCG_01611 [Bacteroides sp. 2_2_4]
Length=381
Score = 52.8 bits (125), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 53/144 (37%), Positives = 79/144 (55%), Gaps = 10/144 (7%)
Query 37 QRIQNEWASSESQKSRDFAKSMFDATNEWNSAKNQRARLEEAGLNPYLMMNggsagtass 96
Q++ ++W+ K A MF+ATNE+NSA QR R E AGLNPY+MMN GSAGTA++
Sbjct 66 QQVSDQWSFYNDAKQN--AWDMFNATNEYNSASAQRERYEAAGLNPYVMMNTGSAGTAAA 123
Query 97 tsantvsgasgsggsPYQFTP-----TNMIGDVASYAAAMKSMSDARKTNTESDLLD--- 148
TSA + + + G +P +P + ++ + + S+ D KT E+ L
Sbjct 124 TSATSATAPTKQGITPPTASPYSADYSGIMQGLGQAIDQLSSIPDKAKTIAETGNLKIEG 183
Query 149 QYGAATYESRIKKTMADTYFTQRQ 172
+Y AA +RI ADT+ + Q
Sbjct 184 KYKAAEAIARIANIKADTHSKKEQ 207
>gi|490418711|ref|WP_004291034.1| hypothetical protein [Bacteroides eggerthii]
gi|217986638|gb|EEC52972.1| hypothetical protein BACEGG_02723 [Bacteroides eggerthii DSM
20697]
Length=368
Score = 50.1 bits (118), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 74/251 (29%), Positives = 103/251 (41%), Gaps = 50/251 (20%)
Query 41 NEWASSESQKSRDFAKSMFDATNEWNSAKNQRARLEEAGLNPYLMMNggsagtasstsan 100
N W E K+ + M++ NE+N QRARLE AGLNPY+MMNGGSAG A S S
Sbjct 77 NAWKLYEDNKA--YQTEMWNKQNEYNDPSAQRARLEAAGLNPYMMMNGGSAGVAGSVSGT 134
Query 101 tvsgasgsggsPYQFTPTNMIGDVASYAAAMKSMSDARKT----------NTESDLL--- 147
S S S P A Y+ M+ + A T N ++D L
Sbjct 135 QGSAPSAGSPSAQGVQPPTATPYSADYSGVMQGLGHAIDTIMTGSQRNIQNAQADNLRIE 194
Query 148 DQYGAA--------TYE---------------SRIKKTMADTYFT-------QRQADVAI 177
+Y A+ TY S I+K ++ + Q QA I
Sbjct 195 GKYIASKAIAELYKTYNEAKNDDERVAIQRVLSSIQKDLSASQVAVNNENVRQIQAQTKI 254
Query 178 AQRANLLLRNDAQEVLNMYLPEEKRIQLQMNGAQYWNMIREGVISEEQAKNLIASRLEIE 237
A NLL + +LP E+R QL + A + ++E+QA++ I E
Sbjct 255 AVTENLLREQQLK-----FLPYEQRTQLALGAADIALKYAQKNLTEKQARHEIEKLAETI 309
Query 238 ARTQGQHISNK 248
R GQ + N+
Sbjct 310 VRANGQAMQNQ 320
>gi|575094319|emb|CDL65706.1| unnamed protein product [uncultured bacterium]
Length=396
Score = 47.8 bits (112), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 76/276 (28%), Positives = 112/276 (41%), Gaps = 51/276 (18%)
Query 31 KHQLEMQRIQNEWASSESQKSRDFAKSMFDATNEWNSAKNQRARLEEAGLNPYLMMNggs 90
K+QL+ R N+ + ++ F + M++ NE+N QRARLE AGLNPYLMM+GGS
Sbjct 50 KYQLQAVRETNQANREIADQNNKFNERMWNLQNEYNRPDMQRARLEAAGLNPYLMMDGGS 109
Query 91 agtasstsantvsgas------gsggsPYQFTPTNMIGDVASYAAAMKSMSDARKTNT-- 142
AG A S SG + YQ N I AS A M D +K N
Sbjct 110 AGIAESAPTADTSGTQIAPDIGNTIAGGYQ-AMGNSISSAASQIAQMTFQDDLQKANVAK 168
Query 143 ------ESDLLDQYGAATYESRIKKTMADTYFTQRQADVAIAQRANLLLRNDAQEVLNM- 195
+ L +Q+ E + + + Q+Q D++ + AN LR+ Q+ L+
Sbjct 169 TVAEAKNAHLQNQFDELRNEFAVANFLVNLRLKQKQGDISDYE-AN-YLRDSMQDRLDSV 226
Query 196 ---------------------------------YLPEEKRIQLQMNGAQYWNMIREGVIS 222
+LP+EK+ L M+ E ++
Sbjct 227 KFQNTLSGSQSSYYSQMAGLTDVQRQIEQTNLDWLPQEKQAGLAATLQNIRTMVSEMGLN 286
Query 223 EEQAKNLIASRLEIEARTQGQHISNKIARSTADAVI 258
QAKN A A +G I N++ ST D +
Sbjct 287 YAQAKNAFAMASLNYANEEGLRIDNRLKESTFDLSV 322
>gi|575094372|emb|CDL65753.1| unnamed protein product [uncultured bacterium]
Length=385
Score = 47.0 bits (110), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 20/44 (45%), Positives = 32/44 (73%), Gaps = 0/44 (0%)
Query 39 IQNEWASSESQKSRDFAKSMFDATNEWNSAKNQRARLEEAGLNP 82
+QNE+ +SE++K+R F KS+++ + WNS NQ + +AGLNP
Sbjct 69 LQNEFNASEAEKNRAFQKSLYERSLSWNSPSNQLKMMADAGLNP 112
>gi|575094301|emb|CDL65691.1| unnamed protein product [uncultured bacterium]
Length=437
Score = 45.8 bits (107), Expect = 0.033, Method: Compositional matrix adjust.
Identities = 23/57 (40%), Positives = 36/57 (63%), Gaps = 3/57 (5%)
Query 31 KHQLEMQRIQNEWASSESQKSRDFAKSMFDATNEWNSAKNQRARLEEAGLNPYLMMN 87
+HQ E R + E+ RDFA+ M+ TN++N+ Q+ RLE+AG+NPY+ M+
Sbjct 45 RHQAEDAR---NFTHQENALQRDFARQMWKDTNDYNTPIAQKQRLEQAGMNPYVNMD 98
>gi|547226428|ref|WP_021963491.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
gi|524103380|emb|CCY83991.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
Length=416
Score = 43.5 bits (101), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 19/22 (86%), Positives = 20/22 (91%), Gaps = 0/22 (0%)
Query 65 WNSAKNQRARLEEAGLNPYLMM 86
+NSAK QRARLE AGLNPYLMM
Sbjct 91 YNSAKAQRARLEAAGLNPYLMM 112
>gi|547836612|ref|WP_022244433.1| putative uncharacterized protein [Roseburia sp. CAG:45]
gi|524471221|emb|CDC12112.1| putative uncharacterized protein [Roseburia sp. CAG:45]
Length=225
Score = 41.6 bits (96), Expect = 0.45, Method: Compositional matrix adjust.
Identities = 20/62 (32%), Positives = 33/62 (53%), Gaps = 0/62 (0%)
Query 281 DVGFRTGKMDRWSQDPVKARWDRGINNAGKFIDGLSNIVGAVTKFGSFRREGRSVIEQFD 340
+V RT MD W+ K R+ + INN+ + D L ++ +T+ ++ R +IE D
Sbjct 161 EVQLRTIAMDFWASLEHKLRYKKHINNSEEIADKLKHVADVITEMDCEMQDIRHMIEFID 220
Query 341 DN 342
DN
Sbjct 221 DN 222
>gi|649555290|gb|KDS61827.1| hypothetical protein M095_3809 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649557306|gb|KDS63785.1| hypothetical protein M095_3404 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649559158|gb|KDS65545.1| hypothetical protein M096_4689 [Parabacteroides distasonis str.
3999B T(B) 6]
gi|649560567|gb|KDS66875.1| hypothetical protein M095_2448 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649561016|gb|KDS67303.1| hypothetical protein M095_2410 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649562727|gb|KDS68911.1| hypothetical protein M096_3341 [Parabacteroides distasonis str.
3999B T(B) 6]
Length=288
Score = 41.6 bits (96), Expect = 0.61, Method: Compositional matrix adjust.
Identities = 20/55 (36%), Positives = 33/55 (60%), Gaps = 0/55 (0%)
Query 31 KHQLEMQRIQNEWASSESQKSRDFAKSMFDATNEWNSAKNQRARLEEAGLNPYLM 85
K +E+ + Q +W E++K+ + M++ NE+NS Q AR+ AGLNP L+
Sbjct 33 KANMEIAKYQAQWQQQENEKAYQRSLKMWNLQNEYNSPTQQMARIRAAGLNPNLV 87
Lambda K H a alpha
0.315 0.127 0.358 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 2103429244710