bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-17_CDS_annotation_glimmer3.pl_2_1
Length=360
Score E
Sequences producing significant alignments: (Bits) Value
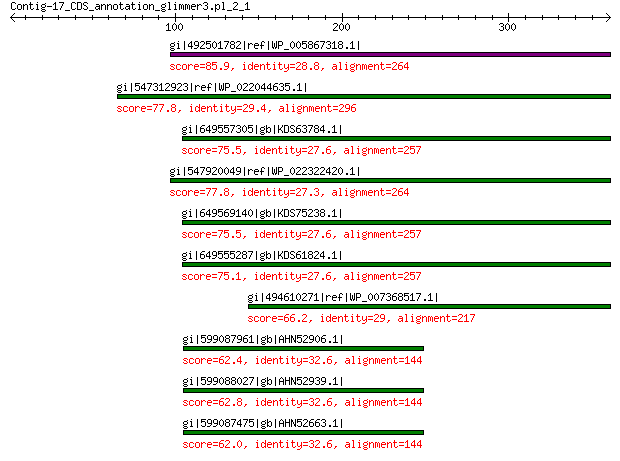
gi|492501782|ref|WP_005867318.1| hypothetical protein 85.9 4e-15
gi|547312923|ref|WP_022044635.1| putative uncharacterized protein 77.8 8e-13
gi|649557305|gb|KDS63784.1| capsid family protein 75.5 2e-12
gi|547920049|ref|WP_022322420.1| capsid protein VP1 77.8 2e-12
gi|649569140|gb|KDS75238.1| capsid family protein 75.5 8e-12
gi|649555287|gb|KDS61824.1| capsid family protein 75.1 1e-11
gi|494610271|ref|WP_007368517.1| capsid protein 66.2 1e-08
gi|599087961|gb|AHN52906.1| major capsid protein 62.4 3e-08
gi|599088027|gb|AHN52939.1| major capsid protein 62.8 3e-08
gi|599087475|gb|AHN52663.1| major capsid protein 62.0 4e-08
>gi|492501782|ref|WP_005867318.1| hypothetical protein [Parabacteroides distasonis]
gi|409230408|gb|EKN23272.1| hypothetical protein HMPREF1059_03257 [Parabacteroides distasonis
CL09T03C24]
Length=538
Score = 85.9 bits (211), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 76/265 (29%), Positives = 118/265 (45%), Gaps = 16/265 (6%)
Query 97 VDVTDGTLSMDALNLSQKVYNFLNRIAVSGGSYRDWLETVYTGGNYMERCETPMFEGGVS 156
V+V + +S++ L S + + R A SG Y + + + + + R + P F GG
Sbjct 289 VNVDELGVSINDLRTSNALQRWFERNARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGR 348
Query 157 QEIVFQEVISNSATEN-EPLGTLAGRGVTTGRQKGGHIRIKITEPCYIMCICSITPRIDY 215
I EV+ SAT++ P +AG G++ G G + E YI+ I SI PR Y
Sbjct 349 TPISVSEVLQTSATDSTSPQANMAGHGISAGVNHG--FKRYFEEHGYIIGIMSIRPRTGY 406
Query 216 GQGNTWDTYLETMDDWHKPALDGIGYQDSLNGERAWWTDYLTADPDLKRTSAGKTVAWIN 275
QG D D++ P +G Q+ N E YL P + G T +
Sbjct 407 QQGVPKDFRKFDNMDFYFPEFAHLGEQEIKNEEV-----YLQQTPASNNGTFGYTPRYAE 461
Query 276 YMTNVNRTFGNFAPGMSESFMVLNRNYSMNNSASPQIEDLTTYIDPVKFNYIFADANIDA 335
Y ++N G+F M +F LNR + S SP + TT+++ N +FA A
Sbjct 462 YKYSMNEVHGDFRGNM--AFWHLNRIF----SESPNLN--TTFVECNPSNRVFATAETSD 513
Query 336 MNFWVQTKFEIKARRLISAKQIPNL 360
+W+Q ++KA RL+ P L
Sbjct 514 DKYWIQLYQDVKALRLMPKYGTPML 538
>gi|547312923|ref|WP_022044635.1| putative uncharacterized protein [Alistipes finegoldii CAG:68]
gi|524208404|emb|CCZ76639.1| putative uncharacterized protein [Alistipes finegoldii CAG:68]
Length=338
Score = 77.8 bits (190), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 87/335 (26%), Positives = 140/335 (42%), Gaps = 48/335 (14%)
Query 65 GLCLKTYNSDLYQNWINTEWIEGVDGINEASAVDV---TDGTLSMDALNLSQKVYNFLNR 121
GL Y+ DL+ N I V+ I +A+D+ T ++++ L L K+ N+++R
Sbjct 11 GLLSVPYSPDLFGNIIKQGSSPAVE-IEVMNALDLNISTGFSVAVPELRLRTKIQNWMDR 69
Query 122 IAVSGGSYRDWLETVYTGGNYMERCETPMFEGGVSQEIVFQEVISNSATENEPLGTLAGR 181
+ VSGG D T++ + P F G V+Q I+ S G+ +G
Sbjct 70 LFVSGGRVGDVFRTLWGTKSSAIYVNKPDFLG------VWQASINPSNVRAMANGSASGE 123
Query 182 GVTTGRQKG---------GH--IRIKITEPCYIMCICSITPRIDYGQGNTWDTYLETMDD 230
G+ GH I EP M I + P Y QG D + D
Sbjct 124 DANLGQLAACVDRYCDFSGHSGIDYYAKEPGTFMLITMLVPEPAYSQGLHPDLASISFGD 183
Query 231 WHKPALDGIGYQ----------------DSLNGERAWWTDY----LTADPDLKRTSAGKT 270
P L+GIG+Q L+ E + W + + DP++ S G+
Sbjct 184 DFNPELNGIGFQLVPRHRFSMMPRGFNFTGLDQEASPWFGHTGTGVLVDPNM--VSVGEE 241
Query 271 VAWINYMTNVNRTFGNFAPGMSESFMVLNRNYSM----NNSASPQIEDLT-TYIDPVKFN 325
VAW T+ +R G+FA + + VL R ++ + + Q + T TYI+P+ +
Sbjct 242 VAWSWLRTDYSRLHGDFAQNGNYQYWVLTRRFTTYFPDDGTGFYQDGEYTGTYINPLDWQ 301
Query 326 YIFADANIDAMNFWVQTKFEIKARRLISAKQIPNL 360
Y+F D + A NF F++ +SA +P L
Sbjct 302 YVFVDQTLMAGNFAYYGTFDLNVTSSLSANYMPYL 336
>gi|649557305|gb|KDS63784.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649559156|gb|KDS65543.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 6]
Length=245
Score = 75.5 bits (184), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 71/258 (28%), Positives = 109/258 (42%), Gaps = 16/258 (6%)
Query 104 LSMDALNLSQKVYNFLNRIAVSGGSYRDWLETVYTGGNYMERCETPMFEGGVSQEIVFQE 163
++++ + S + + R A SG Y + + + + + R + P F GG I E
Sbjct 3 VNINDIRTSNALQRWFERNARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGRTPISVSE 62
Query 164 VISNSATEN-EPLGTLAGRGVTTGRQKGGHIRIKITEPCYIMCICSITPRIDYGQGNTWD 222
V+ S+T++ P +AG G++ G G E YIM I SI PR Y QG D
Sbjct 63 VLQTSSTDSTSPQANMAGHGISAGVNHG--FTRYFEEHGYIMGIMSIRPRTGYQQGVPKD 120
Query 223 TYLETMDDWHKPALDGIGYQDSLNGERAWWTDYLTADPDLKRTSAGKTVAWINYMTNVNR 282
D++ P +G Q+ N E YL + G T + Y + N
Sbjct 121 FRKFDNMDFYFPEFAHLGEQEIKNEEL-----YLNESDAANEGTFGYTPRYAEYKYSQNE 175
Query 283 TFGNFAPGMSESFMVLNRNYSMNNSASPQIEDLTTYIDPVKFNYIFADANIDAMNFWVQT 342
G+F M+ F LNR + P + TT+++ N +FA A +WVQ
Sbjct 176 VHGDFRGNMA--FWHLNRIFK----EKPNLN--TTFVECNPSNRVFATAETSDDKYWVQI 227
Query 343 KFEIKARRLISAKQIPNL 360
+IKA RL+ P L
Sbjct 228 YQDIKALRLMPKYGTPML 245
>gi|547920049|ref|WP_022322420.1| capsid protein VP1 [Parabacteroides merdae CAG:48]
gi|524592961|emb|CDD13573.1| capsid protein VP1 [Parabacteroides merdae CAG:48]
Length=553
Score = 77.8 bits (190), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 72/266 (27%), Positives = 118/266 (44%), Gaps = 18/266 (7%)
Query 97 VDVTDGTLSMDALNLSQKVYNFLNRIAVSGGSYRDWLETVYTGGNYMERCETPMFEGGVS 156
V+V + ++++ L S + + R A G Y + + + + + R + P F GG
Sbjct 304 VNVDEMGININDLRTSNALQRWFERNARGGSRYIEQILSHFGVRSSDARLQRPQFLGGGR 363
Query 157 QEIVFQEVISNSAT-ENEPLGTLAGRGVTTGRQKGGHIRIKITEPCYIMCICSITPRIDY 215
I EV+ S+T E P +AG G++ G G + E YI+ I SITPR Y
Sbjct 364 MPISVSEVLQTSSTDETSPQANMAGHGISAGINNG--FKHYFEEHGYIIGIMSITPRSGY 421
Query 216 GQGNTWD-TYLETMDDWHKPALDGIGYQDSLNGERAWWTDYLTADPDLKRTSAGKTVAWI 274
QG D T + MD ++ P + Q+ N E +++ D + G T +
Sbjct 422 QQGVPRDFTKFDNMD-FYFPEFAHLSEQEIKNQEL-----FVSEDAAYNNGTFGYTPRYA 475
Query 275 NYMTNVNRTFGNFAPGMSESFMVLNRNYSMNNSASPQIEDLTTYIDPVKFNYIFADANID 334
Y + + G+F +S F LNR + P + TT+++ N +FA + +
Sbjct 476 EYKYHPSEAHGDFRGNLS--FWHLNRIFE----DKPNLN--TTFVECKPSNRVFATSETE 527
Query 335 AMNFWVQTKFEIKARRLISAKQIPNL 360
FWVQ ++KA RL+ P L
Sbjct 528 DDKFWVQMYQDVKALRLMPKYGTPML 553
>gi|649569140|gb|KDS75238.1| capsid family protein, partial [Parabacteroides distasonis str.
3999B T(B) 6]
Length=390
Score = 75.5 bits (184), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 71/258 (28%), Positives = 109/258 (42%), Gaps = 16/258 (6%)
Query 104 LSMDALNLSQKVYNFLNRIAVSGGSYRDWLETVYTGGNYMERCETPMFEGGVSQEIVFQE 163
++++ + S + + R A SG Y + + + + + R + P F GG I E
Sbjct 148 VNINDIRTSNALQRWFERNARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGRTPISVSE 207
Query 164 VISNSATEN-EPLGTLAGRGVTTGRQKGGHIRIKITEPCYIMCICSITPRIDYGQGNTWD 222
V+ S+T++ P +AG G++ G G E YIM I SI PR Y QG D
Sbjct 208 VLQTSSTDSTSPQANMAGHGISAGVNHG--FTRYFEEHGYIMGIMSIRPRTGYQQGVPKD 265
Query 223 TYLETMDDWHKPALDGIGYQDSLNGERAWWTDYLTADPDLKRTSAGKTVAWINYMTNVNR 282
D++ P +G Q+ N E YL + G T + Y + N
Sbjct 266 FRKFDNMDFYFPEFAHLGEQEIKNEEL-----YLNESDAANEGTFGYTPRYAEYKYSQNE 320
Query 283 TFGNFAPGMSESFMVLNRNYSMNNSASPQIEDLTTYIDPVKFNYIFADANIDAMNFWVQT 342
G+F M+ F LNR + P + TT+++ N +FA A +WVQ
Sbjct 321 VHGDFRGNMA--FWHLNRIFK----EKPNLN--TTFVECNPSNRVFATAETSDDKYWVQI 372
Query 343 KFEIKARRLISAKQIPNL 360
+IKA RL+ P L
Sbjct 373 YQDIKALRLMPKYGTPML 390
>gi|649555287|gb|KDS61824.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649560568|gb|KDS66876.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649561020|gb|KDS67307.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649562724|gb|KDS68908.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 6]
Length=541
Score = 75.1 bits (183), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 71/258 (28%), Positives = 109/258 (42%), Gaps = 16/258 (6%)
Query 104 LSMDALNLSQKVYNFLNRIAVSGGSYRDWLETVYTGGNYMERCETPMFEGGVSQEIVFQE 163
++++ + S + + R A SG Y + + + + + R + P F GG I E
Sbjct 299 VNINDIRTSNALQRWFERNARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGRTPISVSE 358
Query 164 VISNSATEN-EPLGTLAGRGVTTGRQKGGHIRIKITEPCYIMCICSITPRIDYGQGNTWD 222
V+ S+T++ P +AG G++ G G E YIM I SI PR Y QG D
Sbjct 359 VLQTSSTDSTSPQANMAGHGISAGVNHG--FTRYFEEHGYIMGIMSIRPRTGYQQGVPKD 416
Query 223 TYLETMDDWHKPALDGIGYQDSLNGERAWWTDYLTADPDLKRTSAGKTVAWINYMTNVNR 282
D++ P +G Q+ N E YL + G T + Y + N
Sbjct 417 FRKFDNMDFYFPEFAHLGEQEIKNEEL-----YLNESDAANEGTFGYTPRYAEYKYSQNE 471
Query 283 TFGNFAPGMSESFMVLNRNYSMNNSASPQIEDLTTYIDPVKFNYIFADANIDAMNFWVQT 342
G+F M+ F LNR + P + TT+++ N +FA A +WVQ
Sbjct 472 VHGDFRGNMA--FWHLNRIFK----EKPNLN--TTFVECNPSNRVFATAETSDDKYWVQI 523
Query 343 KFEIKARRLISAKQIPNL 360
+IKA RL+ P L
Sbjct 524 YQDIKALRLMPKYGTPML 541
>gi|494610271|ref|WP_007368517.1| capsid protein [Prevotella multiformis]
gi|324988543|gb|EGC20506.1| putative capsid protein (F protein) [Prevotella multiformis DSM
16608]
Length=531
Score = 66.2 bits (160), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 63/248 (25%), Positives = 106/248 (43%), Gaps = 34/248 (14%)
Query 144 ERCETPMFEGGVSQEIVFQEVISNS-----ATENEPLGTLAGRGVTTGRQKGGHIRIKIT 198
R F GG +V EV++ S A E+ LG L G+GV G I +
Sbjct 287 SRAGDARFIGGFDNPVVISEVVNQSEFDRGADESPCLGDLGGKGV--GSLNSSSIDFDVK 344
Query 199 EPCYIMCICSITPRIDYGQGNTWDTYLETM--DDWHKPALDGIGYQDSLNGE--RAWWTD 254
E IMCI S+ P+ +Y G +D + + +D+ +P +GYQ + + + +
Sbjct 345 EHGIIMCIYSVVPQTEY-NGTYFDPFNRKLRREDFFQPEFADLGYQPVVTSDLISTYLDN 403
Query 255 YLTADPD-LKRTSAGKTVAWIN--------------YMTNVNRTFGNFAPGMSESFMVLN 299
+ P+ KR +AG ++ I Y T+ + FG F G+S S+
Sbjct 404 PVPDGPEKQKRLAAGYPLSSIEANNRLLGWQVRYNEYKTSRDLVFGEFESGLSLSYWCSP 463
Query 300 R-NYSMNNSASPQI------EDLTTYIDPVKFNYIFADANIDAMNFWVQTKFEIKARRLI 352
R ++ + A + Y++P N IF + + A +F V + F++KA R +
Sbjct 464 RYDFGFDGKAGDKKLVNSPWSPAHFYVNPSILNTIFLVSAVKADHFLVNSFFDVKAVRPM 523
Query 353 SAKQIPNL 360
S + L
Sbjct 524 SVSGLAGL 531
>gi|599087961|gb|AHN52906.1| major capsid protein, partial [uncultured Gokushovirinae]
Length=210
Score = 62.4 bits (150), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 47/144 (33%), Positives = 65/144 (45%), Gaps = 3/144 (2%)
Query 105 SMDALNLSQKVYNFLNRIAVSGGSYRDWLETVYTGGNYMERCETPMFEGGVSQEIVFQEV 164
+++ L + ++ L R A SG Y + ++ + G N+M+ P F GG S I V
Sbjct 68 TINQLRQAFQIQKLLERDARSGTRYSEIVKAHF-GVNFMDVTYRPEFLGGTSTPINVTSV 126
Query 165 ISNSATENEPLGTLAGRGVTTGRQKGGHIRIKITEPCYIMCICSITPRIDYGQGNTWDTY 224
S + P GTLA G T GG TE C +M I S+ + Y QG
Sbjct 127 PQTSESGTTPQGTLAAFGTAT--INGGGFTKSFTEHCIVMGIASVRADLTYQQGLNRMFS 184
Query 225 LETMDDWHKPALDGIGYQDSLNGE 248
T D++ PAL IG Q LN E
Sbjct 185 RSTRYDFYFPALAHIGEQSVLNKE 208
>gi|599088027|gb|AHN52939.1| major capsid protein, partial [uncultured Gokushovirinae]
Length=219
Score = 62.8 bits (151), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 47/144 (33%), Positives = 65/144 (45%), Gaps = 3/144 (2%)
Query 105 SMDALNLSQKVYNFLNRIAVSGGSYRDWLETVYTGGNYMERCETPMFEGGVSQEIVFQEV 164
+++ L + ++ L R A SG Y + ++ + G N+M+ P F GG S I V
Sbjct 77 TINQLRQAFQIQKLLERDARSGTRYAEIVKAHF-GVNFMDVTYRPEFLGGTSTPINVTSV 135
Query 165 ISNSATENEPLGTLAGRGVTTGRQKGGHIRIKITEPCYIMCICSITPRIDYGQGNTWDTY 224
S + P GTLA G T GG TE C +M I S+ + Y QG
Sbjct 136 PQTSESGTTPQGTLAAFGTAT--VNGGGFTKSFTEHCIVMGIASVRADLTYQQGLNRMFS 193
Query 225 LETMDDWHKPALDGIGYQDSLNGE 248
T D++ PAL IG Q LN E
Sbjct 194 RSTRYDFYFPALAHIGEQAVLNKE 217
>gi|599087475|gb|AHN52663.1| major capsid protein, partial [uncultured Gokushovirinae]
Length=210
Score = 62.0 bits (149), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 47/144 (33%), Positives = 65/144 (45%), Gaps = 3/144 (2%)
Query 105 SMDALNLSQKVYNFLNRIAVSGGSYRDWLETVYTGGNYMERCETPMFEGGVSQEIVFQEV 164
+++ L + ++ L R A SG Y + ++ + G N+M+ P F GG S I V
Sbjct 68 TINQLRQAFQIQKLLERDARSGTRYSEIVKAHF-GVNFMDVTYRPEFLGGTSTPINVTSV 126
Query 165 ISNSATENEPLGTLAGRGVTTGRQKGGHIRIKITEPCYIMCICSITPRIDYGQGNTWDTY 224
S + P GTLA G T GG TE C +M I S+ + Y QG
Sbjct 127 PQTSESGTTPQGTLAAFGTAT--INGGGFTKSFTEHCILMGIASVRADLTYQQGLNRMFS 184
Query 225 LETMDDWHKPALDGIGYQDSLNGE 248
T D++ PAL IG Q LN E
Sbjct 185 RSTRYDFYFPALAHIGEQSVLNKE 208
Lambda K H a alpha
0.317 0.133 0.407 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 2164993027482