bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-1_CDS_annotation_glimmer3.pl_2_5
Length=113
Score E
Sequences producing significant alignments: (Bits) Value
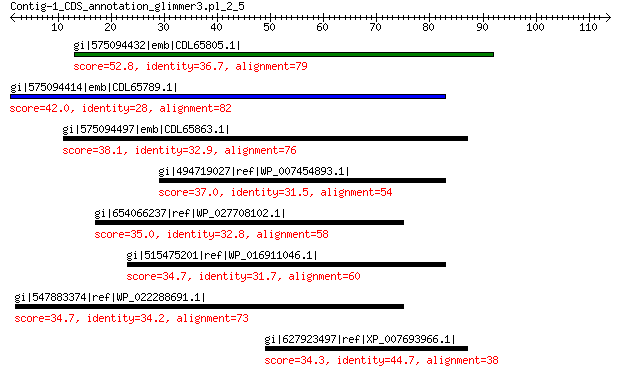
gi|575094432|emb|CDL65805.1| unnamed protein product 52.8 2e-06
gi|575094414|emb|CDL65789.1| unnamed protein product 42.0 0.012
gi|575094497|emb|CDL65863.1| unnamed protein product 38.1 0.44
gi|494719027|ref|WP_007454893.1| amino acid ABC transporter 37.0 1.4
gi|654066237|ref|WP_027708102.1| hypothetical protein 35.0 6.7
gi|515475201|ref|WP_016911046.1| hypothetical protein 34.7 9.1
gi|547883374|ref|WP_022288691.1| efflux transporter RND family M... 34.7 9.1
gi|627923497|ref|XP_007693966.1| hypothetical protein COCMIDRAFT... 34.3 9.7
>gi|575094432|emb|CDL65805.1| unnamed protein product [uncultured bacterium]
Length=158
Score = 52.8 bits (125), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 29/79 (37%), Positives = 46/79 (58%), Gaps = 1/79 (1%)
Query 13 GQEVLQETAPIDIQQEIESYADECDIKSIIRKASFDPQFLKSLSEGAMTGAEVDITEWPQ 72
G++ L+ + +++ +E I++I+R+A+FDP+ L S +E VDIT P
Sbjct 36 GRKELRLVGRESVYDKVQESLEETKIQNIVRRATFDPEILGS-TEWMEKQGMVDITGLPT 94
Query 73 NIHEYHRMIATAQANAMKL 91
NIHEYH + TAQ + KL
Sbjct 95 NIHEYHDFMLTAQKDFDKL 113
>gi|575094414|emb|CDL65789.1| unnamed protein product [uncultured bacterium]
Length=167
Score = 42.0 bits (97), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 23/82 (28%), Positives = 39/82 (48%), Gaps = 5/82 (6%)
Query 1 MRKQYVWTKDEKGQEVLQETAPIDIQQEIESYADECDIKSIIRKASFDPQFLKSLSEGAM 60
+ Y+ E G+ L++ + EI+SY DECD+ SI+ + F S +
Sbjct 24 LEPHYIEKIGENGRTYLEKDGETNTYAEIQSYKDECDVHSILCR-----YFAGDTSVLSR 78
Query 61 TGAEVDITEWPQNIHEYHRMIA 82
G +D T+ P HE + ++A
Sbjct 79 QGVYIDATQLPTTYHEMYNLMA 100
>gi|575094497|emb|CDL65863.1| unnamed protein product [uncultured bacterium]
Length=170
Score = 38.1 bits (87), Expect = 0.44, Method: Compositional matrix adjust.
Identities = 25/77 (32%), Positives = 37/77 (48%), Gaps = 6/77 (8%)
Query 11 EKGQEVLQETAPIDIQQEIESYADECDIKSII-RKASFDPQFLKSLSEGAMTGAEVDITE 69
E G+ L++ ID+ +I+SY D CDI I+ R A D L + G D T
Sbjct 32 EDGKRELEKVGRIDLYAQIQSYKDSCDINYILERFARGDESALSKIQ-----GVYGDFTA 86
Query 70 WPQNIHEYHRMIATAQA 86
P N+ E + + A+A
Sbjct 87 MPTNLAELQQRVVDAEA 103
>gi|494719027|ref|WP_007454893.1| amino acid ABC transporter [Micromonospora lupini]
gi|385888495|emb|CCH15633.1| putative Amino acid ABC transporter [Micromonospora lupini str.
Lupac 08]
Length=315
Score = 37.0 bits (84), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 17/54 (31%), Positives = 31/54 (57%), Gaps = 0/54 (0%)
Query 29 IESYADECDIKSIIRKASFDPQFLKSLSEGAMTGAEVDITEWPQNIHEYHRMIA 82
+++ AD C K ++KAS + L + S+G+ G V++ E+P+N +IA
Sbjct 169 VKTVADLCGRKVAVQKASNQAKNLAAYSKGSCAGRAVEVKEYPENPQAVQALIA 222
>gi|654066237|ref|WP_027708102.1| hypothetical protein [Zooshikella ganghwensis]
Length=334
Score = 35.0 bits (79), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 19/63 (30%), Positives = 34/63 (54%), Gaps = 5/63 (8%)
Query 17 LQETAPIDIQQEIESYADECDIKSIIR-KASFDPQFLKSLSEGAMTGA----EVDITEWP 71
L+E PIDI ++ D D+ +++R K F+P+F+ L G + A +V+ +W
Sbjct 123 LEELPPIDIVIISHNHYDHLDLPTLVRLKKQFNPRFVVGLGNGKLLQAAGITKVNEVDWR 182
Query 72 QNI 74
Q +
Sbjct 183 QTL 185
>gi|515475201|ref|WP_016911046.1| hypothetical protein [Staphylococcus vitulinus]
Length=802
Score = 34.7 bits (78), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 19/60 (32%), Positives = 30/60 (50%), Gaps = 9/60 (15%)
Query 23 IDIQQEIESYADECDIKSIIRKASFDPQFLKSLSEGAMTGAEVDITEWPQNIHEYHRMIA 82
I I +E+ S DE +KS+IR F P+ +T ++++EW N H R+ A
Sbjct 50 IGISEELSSDIDESKLKSLIRVKDFKPE---------LTAELIELSEWMSNFHLVRRISA 100
>gi|547883374|ref|WP_022288691.1| efflux transporter RND family MFP subunit [Ruminococcus sp. CAG:57]
gi|524532065|emb|CDC66858.1| efflux transporter RND family MFP subunit [Ruminococcus sp. CAG:57]
Length=478
Score = 34.7 bits (78), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 25/74 (34%), Positives = 39/74 (53%), Gaps = 5/74 (7%)
Query 2 RKQYVWTKDEKGQEVLQETAPIDIQQEIESYADECDIKSIIRKASFDPQFLKSLSEG-AM 60
+K YVW KD+ G+ E ++I Q+ E D C+IKS + K + KS+ EG
Sbjct 334 KKNYVWAKDKDGK---IEKRYVEIGQKDEDNGD-CEIKSGLEKGDYIAYPDKSIEEGMTA 389
Query 61 TGAEVDITEWPQNI 74
T E D++ P ++
Sbjct 390 TTNEADVSVPPNDL 403
>gi|627923497|ref|XP_007693966.1| hypothetical protein COCMIDRAFT_41947 [Bipolaris oryzae ATCC
44560]
gi|576925581|gb|EUC39516.1| hypothetical protein COCMIDRAFT_41947 [Bipolaris oryzae ATCC
44560]
Length=206
Score = 34.3 bits (77), Expect = 9.7, Method: Compositional matrix adjust.
Identities = 17/39 (44%), Positives = 25/39 (64%), Gaps = 1/39 (3%)
Query 49 PQFLKSLSEGAMTGAEVDITEWPQN-IHEYHRMIATAQA 86
P+FL LS+G +G EV++T+ N I YH + A A+A
Sbjct 65 PEFLTKLSKGQPSGIEVEVTDLETNSISTYHAIKAAARA 103
Lambda K H a alpha
0.313 0.127 0.362 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 434988574272