bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-26_CDS_annotation_glimmer3.pl_2_7
Length=173
Score E
Sequences producing significant alignments: (Bits) Value
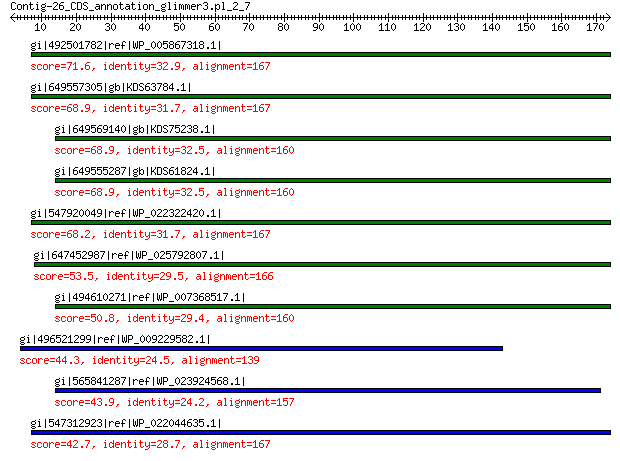
gi|492501782|ref|WP_005867318.1| hypothetical protein 71.6 2e-11
gi|649557305|gb|KDS63784.1| capsid family protein 68.9 2e-11
gi|649569140|gb|KDS75238.1| capsid family protein 68.9 7e-11
gi|649555287|gb|KDS61824.1| capsid family protein 68.9 1e-10
gi|547920049|ref|WP_022322420.1| capsid protein VP1 68.2 2e-10
gi|647452987|ref|WP_025792807.1| hypothetical protein 53.5 2e-05
gi|494610271|ref|WP_007368517.1| capsid protein 50.8 1e-04
gi|496521299|ref|WP_009229582.1| capsid protein 44.3 0.015
gi|565841287|ref|WP_023924568.1| hypothetical protein 43.9 0.022
gi|547312923|ref|WP_022044635.1| putative uncharacterized protein 42.7 0.045
>gi|492501782|ref|WP_005867318.1| hypothetical protein [Parabacteroides distasonis]
gi|409230408|gb|EKN23272.1| hypothetical protein HMPREF1059_03257 [Parabacteroides distasonis
CL09T03C24]
Length=538
Score = 71.6 bits (174), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 55/168 (33%), Positives = 82/168 (49%), Gaps = 13/168 (8%)
Query 7 GLKIKCTEPCMIMALGSITPRIDYSQG-NKWWTRLQNMDDFHKPTLDAIGFQELIaeeaa 65
G K E I+ + SI PR Y QG K + + NMD F+ P +G QE+ EE
Sbjct 383 GFKRYFEEHGYIIGIMSIRPRTGYQQGVPKDFRKFDNMD-FYFPEFAHLGEQEIKNEEVY 441
Query 66 awsteatGNHELVYQSLGKQPSWIEYTTDVNETYGDFAAGMPLAFMCLNRVYEENSDHTI 125
T A+ N + G P + EY +NE +GDF M AF LNR++ E+ +
Sbjct 442 LQQTPASNN-----GTFGYTPRYAEYKYSMNEVHGDFRGNM--AFWHLNRIFSESPNLN- 493
Query 126 ANASTYIDPTIYNKIFAESRLSSQNFWVQVAFDVTARRVMSAKQIPNL 173
+T+++ N++FA + S +W+Q+ DV A R+M P L
Sbjct 494 ---TTFVECNPSNRVFATAETSDDKYWIQLYQDVKALRLMPKYGTPML 538
>gi|649557305|gb|KDS63784.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649559156|gb|KDS65543.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 6]
Length=245
Score = 68.9 bits (167), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 53/168 (32%), Positives = 77/168 (46%), Gaps = 13/168 (8%)
Query 7 GLKIKCTEPCMIMALGSITPRIDYSQG-NKWWTRLQNMDDFHKPTLDAIGFQELIaeeaa 65
G E IM + SI PR Y QG K + + NMD F+ P +G QE I E
Sbjct 90 GFTRYFEEHGYIMGIMSIRPRTGYQQGVPKDFRKFDNMD-FYFPEFAHLGEQE-IKNEEL 147
Query 66 awsteatGNHELVYQSLGKQPSWIEYTTDVNETYGDFAAGMPLAFMCLNRVYEENSDHTI 125
+ N + G P + EY NE +GDF M AF LNR+++E +
Sbjct 148 YLNESDAANE----GTFGYTPRYAEYKYSQNEVHGDFRGNM--AFWHLNRIFKEKPNLN- 200
Query 126 ANASTYIDPTIYNKIFAESRLSSQNFWVQVAFDVTARRVMSAKQIPNL 173
+T+++ N++FA + S +WVQ+ D+ A R+M P L
Sbjct 201 ---TTFVECNPSNRVFATAETSDDKYWVQIYQDIKALRLMPKYGTPML 245
>gi|649569140|gb|KDS75238.1| capsid family protein, partial [Parabacteroides distasonis str.
3999B T(B) 6]
Length=390
Score = 68.9 bits (167), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 52/161 (32%), Positives = 76/161 (47%), Gaps = 13/161 (8%)
Query 14 EPCMIMALGSITPRIDYSQG-NKWWTRLQNMDDFHKPTLDAIGFQELIaeeaaawsteat 72
E IM + SI PR Y QG K + + NMD F+ P +G QE I E +
Sbjct 242 EHGYIMGIMSIRPRTGYQQGVPKDFRKFDNMD-FYFPEFAHLGEQE-IKNEELYLNESDA 299
Query 73 GNHELVYQSLGKQPSWIEYTTDVNETYGDFAAGMPLAFMCLNRVYEENSDHTIANASTYI 132
N + G P + EY NE +GDF M AF LNR+++E + +T++
Sbjct 300 ANE----GTFGYTPRYAEYKYSQNEVHGDFRGNM--AFWHLNRIFKEKPNLN----TTFV 349
Query 133 DPTIYNKIFAESRLSSQNFWVQVAFDVTARRVMSAKQIPNL 173
+ N++FA + S +WVQ+ D+ A R+M P L
Sbjct 350 ECNPSNRVFATAETSDDKYWVQIYQDIKALRLMPKYGTPML 390
>gi|649555287|gb|KDS61824.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649560568|gb|KDS66876.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649561020|gb|KDS67307.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649562724|gb|KDS68908.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 6]
Length=541
Score = 68.9 bits (167), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 52/161 (32%), Positives = 76/161 (47%), Gaps = 13/161 (8%)
Query 14 EPCMIMALGSITPRIDYSQG-NKWWTRLQNMDDFHKPTLDAIGFQELIaeeaaawsteat 72
E IM + SI PR Y QG K + + NMD F+ P +G QE I E +
Sbjct 393 EHGYIMGIMSIRPRTGYQQGVPKDFRKFDNMD-FYFPEFAHLGEQE-IKNEELYLNESDA 450
Query 73 GNHELVYQSLGKQPSWIEYTTDVNETYGDFAAGMPLAFMCLNRVYEENSDHTIANASTYI 132
N + G P + EY NE +GDF M AF LNR+++E + +T++
Sbjct 451 ANE----GTFGYTPRYAEYKYSQNEVHGDFRGNM--AFWHLNRIFKEKPNLN----TTFV 500
Query 133 DPTIYNKIFAESRLSSQNFWVQVAFDVTARRVMSAKQIPNL 173
+ N++FA + S +WVQ+ D+ A R+M P L
Sbjct 501 ECNPSNRVFATAETSDDKYWVQIYQDIKALRLMPKYGTPML 541
>gi|547920049|ref|WP_022322420.1| capsid protein VP1 [Parabacteroides merdae CAG:48]
gi|524592961|emb|CDD13573.1| capsid protein VP1 [Parabacteroides merdae CAG:48]
Length=553
Score = 68.2 bits (165), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 53/168 (32%), Positives = 79/168 (47%), Gaps = 13/168 (8%)
Query 7 GLKIKCTEPCMIMALGSITPRIDYSQG-NKWWTRLQNMDDFHKPTLDAIGFQELIaeeaa 65
G K E I+ + SITPR Y QG + +T+ NMD F+ P + QE+ +E
Sbjct 398 GFKHYFEEHGYIIGIMSITPRSGYQQGVPRDFTKFDNMD-FYFPEFAHLSEQEIKNQELF 456
Query 66 awsteatGNHELVYQSLGKQPSWIEYTTDVNETYGDFAAGMPLAFMCLNRVYEENSDHTI 125
A N + G P + EY +E +GDF L+F LNR++E+ +
Sbjct 457 VSEDAAYNN-----GTFGYTPRYAEYKYHPSEAHGDFRGN--LSFWHLNRIFEDKPNLN- 508
Query 126 ANASTYIDPTIYNKIFAESRLSSQNFWVQVAFDVTARRVMSAKQIPNL 173
+T+++ N++FA S FWVQ+ DV A R+M P L
Sbjct 509 ---TTFVECKPSNRVFATSETEDDKFWVQMYQDVKALRLMPKYGTPML 553
>gi|647452987|ref|WP_025792807.1| hypothetical protein [Prevotella histicola]
Length=584
Score = 53.5 bits (127), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 49/198 (25%), Positives = 83/198 (42%), Gaps = 36/198 (18%)
Query 8 LKIKCTEPCMIMALGSITPRIDYSQG-----NKWWTRLQNMDDFHKPTLDAIGFQELI-- 60
++ TE +IM + S+ P+ +Y+ N+ TR Q F++P +G+Q LI
Sbjct 391 IEFDSTEHGIIMCIYSVAPQSEYNASYLDPFNRKLTREQ----FYQPEFADLGYQALIGS 446
Query 61 ----aeeaaawsteatGNHELVYQSLGKQPSWIEYTTDVNETYGDFAAGMPLAFMCLNRV 116
+ + EL LG Q + EY T + +GDF +G L++ C R
Sbjct 447 DLICSTLGMNEKQAGFSDIELNNNLLGYQVRYNEYKTARDLVFGDFESGKSLSYWCTPRF 506
Query 117 --------------------YEENSDHTI-ANASTYIDPTIYNKIFAESRLSSQNFWVQV 155
Y + + + ++ + YI+P + N IF S + + +F V
Sbjct 507 DFGYGDTEKKIAPENKGGADYRKKGNRSHWSSRNFYINPNLVNPIFLTSAVQADHFIVNS 566
Query 156 AFDVTARRVMSAKQIPNL 173
DV A R MS + +L
Sbjct 567 FLDVKAVRPMSVTGLSSL 584
>gi|494610271|ref|WP_007368517.1| capsid protein [Prevotella multiformis]
gi|324988543|gb|EGC20506.1| putative capsid protein (F protein) [Prevotella multiformis DSM
16608]
Length=531
Score = 50.8 bits (120), Expect = 1e-04, Method: Composition-based stats.
Identities = 47/189 (25%), Positives = 84/189 (44%), Gaps = 31/189 (16%)
Query 14 EPCMIMALGSITPRIDYSQGNKW--WTRLQNMDDFHKPTLDAIGFQELIaeeaaawstea 71
E +IM + S+ P+ +Y+ G + + R +DF +P +G+Q ++ + + +
Sbjct 345 EHGIIMCIYSVVPQTEYN-GTYFDPFNRKLRREDFFQPEFADLGYQPVVTSDLISTYLDN 403
Query 72 -----------------tGNHELVYQSLGKQPSWIEYTTDVNETYGDFAAGMPLAFMCLN 114
+ E + LG Q + EY T + +G+F +G+ L++ C
Sbjct 404 PVPDGPEKQKRLAAGYPLSSIEANNRLLGWQVRYNEYKTSRDLVFGEFESGLSLSYWCSP 463
Query 115 RVYE-----ENSDHTIAN-----ASTYIDPTIYNKIFAESRLSSQNFWVQVAFDVTARRV 164
R Y+ + D + N A Y++P+I N IF S + + +F V FDV A R
Sbjct 464 R-YDFGFDGKAGDKKLVNSPWSPAHFYVNPSILNTIFLVSAVKADHFLVNSFFDVKAVRP 522
Query 165 MSAKQIPNL 173
MS + L
Sbjct 523 MSVSGLAGL 531
>gi|496521299|ref|WP_009229582.1| capsid protein [Prevotella sp. oral taxon 317]
gi|288330570|gb|EFC69154.1| putative capsid protein (F protein) [Prevotella sp. oral taxon
317 str. F0108]
Length=541
Score = 44.3 bits (103), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 34/141 (24%), Positives = 62/141 (44%), Gaps = 10/141 (7%)
Query 4 SGRG-LKIKCTEPCMIMALGSITPRIDYS-QGNKWWTRLQNMDDFHKPTLDAIGFQELIa 61
SG G ++ EP ++M + S+ P + Y + Q D+ P + +G Q ++
Sbjct 375 SGYGEIQFDAKEPGVLMCIYSVVPAMQYDCMRLDPFVAKQTRGDYFIPEFENLGMQPIVP 434
Query 62 eeaaawsteatGNHELVYQSLGKQPSWIEYTTDVNETYGDFAAGMPLAFMCLNRVYEENS 121
+ + S G QP + EY T + +G FA G PL++ + R ++
Sbjct 435 AFVSLNRAKD--------NSYGWQPRYSEYKTAFDINHGQFANGEPLSYWSIARARGSDT 486
Query 122 DHTIANASTYIDPTIYNKIFA 142
+T A+ I+P + +FA
Sbjct 487 LNTFNVAALKINPHWLDSVFA 507
>gi|565841287|ref|WP_023924568.1| hypothetical protein [Prevotella nigrescens]
gi|564729907|gb|ETD29851.1| hypothetical protein HMPREF1173_00033 [Prevotella nigrescens
CC14M]
Length=656
Score = 43.9 bits (102), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 38/162 (23%), Positives = 71/162 (44%), Gaps = 5/162 (3%)
Query 14 EPCMIMALGSITPRIDY-SQGNKWWTRLQNMDDFHKPTLDAIGFQELIaeeaaawsteat 72
E +IM + SI P++DY ++ + R + +D+ +P + +G Q +I + A
Sbjct 492 EHGLIMCIYSIAPQVDYDARELDPFNRKFSREDYFQPEFENLGMQPVIQSDLCLCINSAK 551
Query 73 GNHELVYQS-LGKQPSWIEYTTDVNETYGDFAAGMPLAFMCLNRVYEENSDHTIANASTY 131
+ + + LG ++EY T + +G+F +G L+ + ++
Sbjct 552 SDSSDQHNNVLGYSARYLEYKTARDIIFGEFMSGGSLSAWATPKNNYTFEFGKLSLPDLL 611
Query 132 IDPTIYNKIFA---ESRLSSQNFWVQVAFDVTARRVMSAKQI 170
+DP + IFA +S+ F V FDV A R M +
Sbjct 612 VDPKVLEPIFAVKYNGSMSTDQFLVNSYFDVKAIRPMQVNDM 653
>gi|547312923|ref|WP_022044635.1| putative uncharacterized protein [Alistipes finegoldii CAG:68]
gi|524208404|emb|CCZ76639.1| putative uncharacterized protein [Alistipes finegoldii CAG:68]
Length=338
Score = 42.7 bits (99), Expect = 0.045, Method: Compositional matrix adjust.
Identities = 48/197 (24%), Positives = 77/197 (39%), Gaps = 35/197 (18%)
Query 7 GLKIKCTEPCMIMALGSITPRIDYSQG-NKWWTRLQNMDDFHKPTLDAIGFQELIaeeaa 65
G+ EP M + + P YSQG + + DDF+ P L+ IGFQ + +
Sbjct 145 GIDYYAKEPGTFMLITMLVPEPAYSQGLHPDLASISFGDDFN-PELNGIGFQLVPRHRFS 203
Query 66 awste---------------atGNHELV---YQSLGKQPSWIEYTTDVNETYGDFAAGMP 107
TG LV S+G++ +W TD + +GDFA
Sbjct 204 MMPRGFNFTGLDQEASPWFGHTGTGVLVDPNMVSVGEEVAWSWLRTDYSRLHGDFAQNGN 263
Query 108 LAFMCLNRVYE-----------ENSDHTIANASTYIDPTIYNKIFAESRLSSQNFWVQVA 156
+ L R + ++ ++T TYI+P + +F + L + NF
Sbjct 264 YQYWVLTRRFTTYFPDDGTGFYQDGEYT----GTYINPLDWQYVFVDQTLMAGNFAYYGT 319
Query 157 FDVTARRVMSAKQIPNL 173
FD+ +SA +P L
Sbjct 320 FDLNVTSSLSANYMPYL 336
Lambda K H a alpha
0.320 0.134 0.416 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 428836147623