bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-28_CDS_annotation_glimmer3.pl_2_1
Length=324
Score E
Sequences producing significant alignments: (Bits) Value
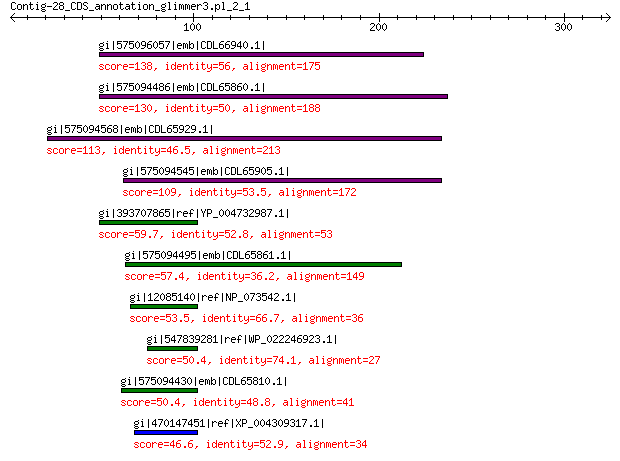
gi|575096057|emb|CDL66940.1| unnamed protein product 138 1e-34
gi|575094486|emb|CDL65860.1| unnamed protein product 130 3e-31
gi|575094568|emb|CDL65929.1| unnamed protein product 113 2e-25
gi|575094545|emb|CDL65905.1| unnamed protein product 109 6e-24
gi|393707865|ref|YP_004732987.1| structural protein VP2 59.7 3e-07
gi|575094495|emb|CDL65861.1| unnamed protein product 57.4 3e-06
gi|12085140|ref|NP_073542.1| minor capsid protein 53.5 3e-05
gi|547839281|ref|WP_022246923.1| putative minor capsid protein 50.4 5e-04
gi|575094430|emb|CDL65810.1| unnamed protein product 50.4 6e-04
gi|470147451|ref|XP_004309317.1| PREDICTED: minor spike protein ... 46.6 0.003
>gi|575096057|emb|CDL66940.1| unnamed protein product [uncultured bacterium]
Length=275
Score = 138 bits (347), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 98/175 (56%), Positives = 122/175 (70%), Gaps = 0/175 (0%)
Query 49 EKQMDFQREQNKAAMDFNAAEAAKNRDWQKMMSDTAHQREIRDLQAAGLNPILsatggng 108
E Q D+Q QN AM FN+ EAAKNR WQ+MMS+TAHQRE++DL AAGLNP+LSA GNG
Sbjct 44 EIQRDWQEAQNAKAMQFNSMEAAKNRKWQEMMSNTAHQREVKDLMAAGLNPVLSAMNGNG 103
Query 109 aavtsgatasgvtssgaMGQTDTSKNSAIVQLLSSMLQYKNNLDMANINAQTNLAVADKY 168
AAV SGATASGVTS+GA G+ DTS + AI LL S+L + AN+NA+T AVADKY
Sbjct 104 AAVGSGATASGVTSAGAKGEADTSTSGAIANLLGSILSASTAIQAANVNARTQEAVADKY 163
Query 169 NAMSKYMSELNAATNLQTAGMSAAASRDVAGINASASRYASDVHLQAQQYASDIQ 223
AMS+ ++E+N A L +AG+ A A+R A AS + S A +YA+D Q
Sbjct 164 TAMSQIVAEINKAATLGSAGIHAGATRYAADAAKVASIFGSSAAASASRYATDQQ 218
>gi|575094486|emb|CDL65860.1| unnamed protein product [uncultured bacterium]
Length=344
Score = 130 bits (327), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 94/189 (50%), Positives = 127/189 (67%), Gaps = 1/189 (1%)
Query 49 EKQMDFQREQNKAAMDFNAAEAAKNRDWQKMMSDTAHQREIRDLQAAGLNPILsatggng 108
+KQMDFQ Q FN EA +R WQ+ MS+TAHQREI+DLQAAGLNP+LSA GG+G
Sbjct 84 QKQMDFQASQGALVRQFNHDEAELSRLWQERMSNTAHQREIKDLQAAGLNPVLSAMGGSG 143
Query 109 aavtsgatasg-vtssgaMGQTDTSKNSAIVQLLSSMLQYKNNLDMANINAQTNLAVADK 167
A VTSG+TASG SG+ G TDTS A+V LL S + + ++ ++A+T +VADK
Sbjct 144 APVTSGSTASGYSPPSGSKGDTDTSLAGALVSLLGSSMMAQASMANTAMSARTQESVADK 203
Query 168 YNAMSKYMSELNAATNLQTAGMSAAASRDVAGINASASRYASDVHLQAQQYASDIQSYTS 227
Y AMSK ++E+ T L + +SA ASR A +A AS+ A+ +H AQ+Y D+Q+ T
Sbjct 204 YTAMSKLVAEIQQETTLSASTISAMASRYAADRSADASKVAASIHAAAQRYGYDVQAMTQ 263
Query 228 KVVASINAE 236
+ +AS NA+
Sbjct 264 RDIASFNAQ 272
>gi|575094568|emb|CDL65929.1| unnamed protein product [uncultured bacterium]
Length=310
Score = 113 bits (283), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 99/224 (44%), Positives = 143/224 (64%), Gaps = 13/224 (6%)
Query 21 NAVYQANQAFDQInattnannawsaaqaEKQMDFQREQNKAAMDFNAAEAAKNRDWQKMM 80
N + + Q+ D + + N A S +AE +Q EQN+ AM FNAAEA KNR+WQ++M
Sbjct 34 NQLTKITQSIDDVIGLSEKNTARSVQEAESLRTWQEEQNRIAMQFNAAEAEKNRNWQEIM 93
Query 81 SDTAHQREIRDLQAAGLNPILsatggngaavtsgatasgvtssgaMGQTDTSKNSAIVQL 140
S+TAHQRE+ DL AAGLNP+LSA GGNGAAVTSGATASGVTSSGA G DTS +SA+V +
Sbjct 94 SNTAHQREVNDLMAAGLNPVLSAGGGNGAAVTSGATASGVTSSGAKGDVDTSASSAVVGI 153
Query 141 LSSMLQYKNNLDMANINAQTNLAVADKYNAMSKYMSELNAATNLQTA------GMSAAA- 193
L SML N+ AN +A T++A +K +++ ++ N L+ A G++ AA
Sbjct 154 LGSMLSSLTNIANANTSAITSMANTEKLGQINQLIAHANNENALKVAETYGKYGVAQAAT 213
Query 194 ----SRDVAGINASASRYASDVHLQAQQYASDIQSYTSKVVASI 233
S + A ++A A+RY++D + ++Y ++Y + V ++
Sbjct 214 AGRYSLNAAQVHADATRYSADAYTNMEKYLR--ENYPTSTVGAV 255
>gi|575094545|emb|CDL65905.1| unnamed protein product [uncultured bacterium]
Length=325
Score = 109 bits (272), Expect = 6e-24, Method: Compositional matrix adjust.
Identities = 92/172 (53%), Positives = 119/172 (69%), Gaps = 1/172 (1%)
Query 62 AMDFNAAEAAKNRDWQKMMSDTAHQREIRDLQAAGLNPILsatggngaavtsgatasgvt 121
AM F++AEAAKNRDWQ MS+TAHQRE+ DL+AAGLNP+LSA GGNGAAVTSGATA G T
Sbjct 56 AMQFSSAEAAKNRDWQSYMSNTAHQREVADLKAAGLNPVLSAMGGNGAAVTSGATAQGYT 115
Query 122 ssgaMGQTDTSKNSAIVQLLSSMLQYKNNLDMANINAQTNLAVADKYNAMSKYMSELNAA 181
SSG DTS +A+V LL S+L + ++ NA NL+VADKY + ++Y +++ A
Sbjct 116 SSGGQASADTSATAALVGLLGSLLNAQTSIANTATNAVANLSVADKYTSATRYAADVGYA 175
Query 182 TNLQTAGMSAAASRDVAGINASASRYASDVHLQAQQYASDIQSYTSKVVASI 233
+A ++A ASR + +AS+YASD A +YASD QSY + ASI
Sbjct 176 GTSYSANVAAYASRFASNNALAASKYASDNSRAASKYASD-QSYLASKFASI 226
>gi|393707865|ref|YP_004732987.1| structural protein VP2 [Microviridae phi-CA82]
gi|311336637|gb|ADP89808.1| structural protein VP2 [Microviridae phi-CA82]
Length=234
Score = 59.7 bits (143), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 28/53 (53%), Positives = 38/53 (72%), Gaps = 0/53 (0%)
Query 49 EKQMDFQREQNKAAMDFNAAEAAKNRDWQKMMSDTAHQREIRDLQAAGLNPIL 101
+KQ + EQ + + FNA EA KNRDWQ+ MS+TA QR+++D + AGLNPI
Sbjct 13 DKQNKWNAEQTEKSNQFNAQEAQKNRDWQEQMSNTALQRKMQDAEKAGLNPIF 65
>gi|575094495|emb|CDL65861.1| unnamed protein product [uncultured bacterium]
Length=266
Score = 57.4 bits (137), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 54/157 (34%), Positives = 79/157 (50%), Gaps = 30/157 (19%)
Query 63 MDFNAAEAAKNRDWQKMMSDTAHQREIRDLQAAGLNPILsatggngaavtsgatasgvts 122
+++N A + +Q+ MS TAHQRE++DL AAGLNP+LSA
Sbjct 64 LNYNTQSAREQMAFQERMSSTAHQREVKDLIAAGLNPVLSAG------------------ 105
Query 123 sgaMGQTDTSKNSAIVQLLSSMLQYKNN--LDMANINAQ------TNLAVADKYNAMSKY 174
G ++ + A+ SSM+ K N L +NAQ N A D AM+KY
Sbjct 106 ----GSGASAPSGAMATADSSMMSAKANAALQKRIVNAQLKNAKDINKAQLDAQKAMNKY 161
Query 175 MSELNAATNLQTAGMSAAASRDVAGINASASRYASDV 211
++ A T+L A +SA+AS+ A A+AS Y S++
Sbjct 162 SVDVGAQTSLANAQISASASKFGAMQAAAASMYGSNL 198
>gi|12085140|ref|NP_073542.1| minor capsid protein [Bdellovibrio phage phiMH2K]
gi|75089169|sp|Q9G055.1|H_BPPHM RecName: Full=Minor spike protein H; AltName: Full=H protein;
AltName: Full=Pilot protein; AltName: Full=Protein VP2; Short=VP2
[Bdellovibrio phage phiMH2K]
gi|12017988|gb|AAG45344.1|AF306496_5 Vp2 [Bdellovibrio phage phiMH2K]
Length=199
Score = 53.5 bits (127), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 24/36 (67%), Positives = 29/36 (81%), Gaps = 0/36 (0%)
Query 66 NAAEAAKNRDWQKMMSDTAHQREIRDLQAAGLNPIL 101
N AEAA+NR WQ+ MS++AHQRE DLQ AGLN +L
Sbjct 42 NQAEAARNRKWQEQMSNSAHQREANDLQTAGLNRLL 77
>gi|547839281|ref|WP_022246923.1| putative minor capsid protein [Clostridium sp. CAG:306]
gi|524476581|emb|CDC18646.1| putative minor capsid protein [Clostridium sp. CAG:306]
Length=236
Score = 50.4 bits (119), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 20/27 (74%), Positives = 25/27 (93%), Gaps = 0/27 (0%)
Query 75 DWQKMMSDTAHQREIRDLQAAGLNPIL 101
D+Q+ MS TAHQRE++DL+AAGLNPIL
Sbjct 40 DFQERMSSTAHQREVKDLRAAGLNPIL 66
>gi|575094430|emb|CDL65810.1| unnamed protein product [uncultured bacterium]
Length=274
Score = 50.4 bits (119), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 20/41 (49%), Positives = 32/41 (78%), Gaps = 0/41 (0%)
Query 61 AAMDFNAAEAAKNRDWQKMMSDTAHQREIRDLQAAGLNPIL 101
AAM +N+A+A + W++ MS+T++QR + D++ AGLNPIL
Sbjct 119 AAMQYNSAQAMRQMKWEEHMSNTSYQRAMEDMRKAGLNPIL 159
>gi|470147451|ref|XP_004309317.1| PREDICTED: minor spike protein H-like, partial [Fragaria vesca
subsp. vesca]
Length=139
Score = 46.6 bits (109), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 18/34 (53%), Positives = 30/34 (88%), Gaps = 0/34 (0%)
Query 68 AEAAKNRDWQKMMSDTAHQREIRDLQAAGLNPIL 101
AEA +NR++Q+ +S++A+QR++ DL +AGLNP+L
Sbjct 24 AEAQRNREFQERLSNSAYQRQVADLSSAGLNPML 57
Lambda K H a alpha
0.309 0.117 0.308 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1814496934800