bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-28_CDS_annotation_glimmer3.pl_2_3
Length=126
Score E
Sequences producing significant alignments: (Bits) Value
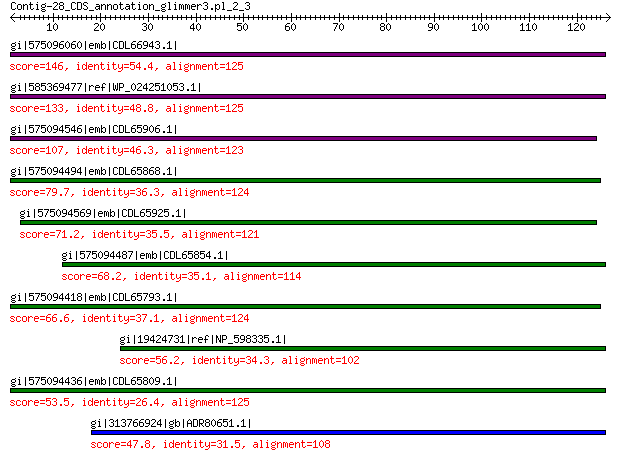
gi|575096060|emb|CDL66943.1| unnamed protein product 146 1e-39
gi|585369477|ref|WP_024251053.1| hypothetical protein 133 8e-36
gi|575094546|emb|CDL65906.1| unnamed protein product 107 3e-25
gi|575094494|emb|CDL65868.1| unnamed protein product 79.7 3e-15
gi|575094569|emb|CDL65925.1| unnamed protein product 71.2 3e-12
gi|575094487|emb|CDL65854.1| unnamed protein product 68.2 4e-11
gi|575094418|emb|CDL65793.1| unnamed protein product 66.6 2e-10
gi|19424731|ref|NP_598335.1| hypothetical protein Sp-4p1 56.2 5e-07
gi|575094436|emb|CDL65809.1| unnamed protein product 53.5 5e-06
gi|313766924|gb|ADR80651.1| putative replication initiation protein 47.8 4e-04
>gi|575096060|emb|CDL66943.1| unnamed protein product [uncultured bacterium]
Length=339
Score = 146 bits (368), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 68/125 (54%), Positives = 92/125 (74%), Gaps = 0/125 (0%)
Query 1 VTKKLTGPAACFYDDHNIEPPFTQMSLKPGIGRNYYDNHPDLYKYEYINVSTAKGGRKFR 60
V KK G A+ Y + N+EP FT MS KPGI RNYYD HPDL++ ++IN+ST KGGRKFR
Sbjct 214 VLKKQKGEASTVYQEFNLEPEFTLMSRKPGIARNYYDTHPDLFQSDFINISTLKGGRKFR 273
Query 61 PPKYFDRIFDVEYPDESAAMKEARKRIAMDARQAKLAQTNLSYIDLLAVEERALKERIKS 120
PP+YF+++F++++P+E+A E RK +A AKLA+TNL + +LAVEER +RIK
Sbjct 274 PPRYFEKLFELDFPEEAAKRSEVRKAAGSNAMAAKLAKTNLDPLSMLAVEERNFTDRIKP 333
Query 121 LKRRL 125
L+R L
Sbjct 334 LRRNL 338
>gi|585369477|ref|WP_024251053.1| hypothetical protein [Escherichia coli]
Length=243
Score = 133 bits (335), Expect = 8e-36, Method: Compositional matrix adjust.
Identities = 61/125 (49%), Positives = 87/125 (70%), Gaps = 0/125 (0%)
Query 1 VTKKLTGPAACFYDDHNIEPPFTQMSLKPGIGRNYYDNHPDLYKYEYINVSTAKGGRKFR 60
V KK GP A Y NI+P + MS +PGIGR +YD+HP+ +Y+ I++ST GGRK R
Sbjct 119 VLKKACGPEADVYQTFNIQPEYVDMSRRPGIGRQWYDDHPECMEYDTISISTPDGGRKIR 178
Query 61 PPKYFDRIFDVEYPDESAAMKEARKRIAMDARQAKLAQTNLSYIDLLAVEERALKERIKS 120
PPKYFD++FD+E P+ A +K RK A + ++AKLAQ+ ++Y ++L +ER L RIK+
Sbjct 179 PPKYFDKLFDLEQPELMAEIKAKRKHFAEEGKKAKLAQSTMTYEEILETQERVLHNRIKN 238
Query 121 LKRRL 125
L+R L
Sbjct 239 LRREL 243
>gi|575094546|emb|CDL65906.1| unnamed protein product [uncultured bacterium]
Length=351
Score = 107 bits (267), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 57/124 (46%), Positives = 75/124 (60%), Gaps = 1/124 (1%)
Query 1 VTKKLTGPAACFYDDHNIEPPFTQMSLKPGIGRNYYDNHP-DLYKYEYINVSTAKGGRKF 59
VTKKLTG A FY N+ PPF+ MS KPGI YY +H D+Y + IN+ST +GG
Sbjct 225 VTKKLTGNLAQFYTTFNLTPPFSLMSRKPGIAYQYYADHGLDIYDNDKINISTERGGLSM 284
Query 60 RPPKYFDRIFDVEYPDESAAMKEARKRIAMDARQAKLAQTNLSYIDLLAVEERALKERIK 119
PPKYFD ++++ PD+ K R +A ++ KL Q+ LSY +LAV E +IK
Sbjct 285 LPPKYFDHFYELDAPDDYVNYKSVRAALARESLLLKLDQSTLSYGQMLAVAESIKNNKIK 344
Query 120 SLKR 123
SL R
Sbjct 345 SLDR 348
>gi|575094494|emb|CDL65868.1| unnamed protein product [uncultured bacterium]
Length=348
Score = 79.7 bits (195), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 45/125 (36%), Positives = 68/125 (54%), Gaps = 1/125 (1%)
Query 1 VTKKLTGPAACFYDDHNIEPPFTQMSLKPGIGRNYY-DNHPDLYKYEYINVSTAKGGRKF 59
+ KK G A Y D+NI P FT MS KP I R YY DN ++ +YI + T + +
Sbjct 222 ILKKQYGSGAQIYKDYNILPEFTCMSTKPAIAREYYEDNKDKIFDSDYIFLGTKEKSIQM 281
Query 60 RPPKYFDRIFDVEYPDESAAMKEARKRIAMDARQAKLAQTNLSYIDLLAVEERALKERIK 119
+PPKYF+++ + E D ++ +A D + T+ Y+ +L +EE L RIK
Sbjct 282 KPPKYFEKLLEKENEDVFKERRDLHASLAEDFSCLRNLSTSHDYLGMLQMEEDNLNARIK 341
Query 120 SLKRR 124
+LKR+
Sbjct 342 TLKRK 346
>gi|575094569|emb|CDL65925.1| unnamed protein product [uncultured bacterium]
Length=354
Score = 71.2 bits (173), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 43/122 (35%), Positives = 67/122 (55%), Gaps = 1/122 (1%)
Query 3 KKLTGPAACFYDDHNIEPPFTQMSLKPGIGRNYYDNHPD-LYKYEYINVSTAKGGRKFRP 61
KK TG + Y+ + P F MS KPGI R Y+D+H D + KY+ IN+ST KGG +
Sbjct 229 KKATGFDSEIYERLGVLPEFVTMSRKPGIARKYFDDHYDEIIKYKTINLSTLKGGMSMQI 288
Query 62 PKYFDRIFDVEYPDESAAMKEARKRIAMDARQAKLAQTNLSYIDLLAVEERALKERIKSL 121
P YF R+ + + +K + K+ A++ ++A + T++ YI L+ E L K
Sbjct 289 PPYFIRLIEDIDSELFKEIKRSNKQAALNHQEALMKNTDVDYITYLSFLEGILVREEKFY 348
Query 122 KR 123
+R
Sbjct 349 RR 350
>gi|575094487|emb|CDL65854.1| unnamed protein product [uncultured bacterium]
Length=332
Score = 68.2 bits (165), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 40/115 (35%), Positives = 62/115 (54%), Gaps = 2/115 (2%)
Query 12 FYDDHNIEPPFTQMSLKPGIGRNYYDNHPDLYKYEYINVSTAKGGRKFRPPKYFDRIFDV 71
Y ++P F MS +PG+ R YY++HPD+++Y NVST +GGRK P +YF +++
Sbjct 218 LYKKAALQPEFQVMSNRPGLARQYYEDHPDIFRYLSFNVSTPQGGRKMYPSEYFRKLYRD 277
Query 72 EYPDESAAMKEARKRIAMDAR-QAKLAQTNLSYIDLLAVEERALKERIKSLKRRL 125
+ E + R R ++ K T+LSY D+L +E R+ L R L
Sbjct 278 GHERELFE-RSLRTREELEVENHLKNMLTDLSYDDILKEDEEREFRRLSHLHRDL 331
>gi|575094418|emb|CDL65793.1| unnamed protein product [uncultured bacterium]
Length=367
Score = 66.6 bits (161), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 46/152 (30%), Positives = 73/152 (48%), Gaps = 31/152 (20%)
Query 1 VTKKLTGPAACFYDDHNIEPPFTQMSLKPGIGRNYYDNHPD-LYKYEYINVSTAKGGRKF 59
V KK G A Y D I P F QMSLKP IG+ Y++ H D +Y + IN+++ GR
Sbjct 214 VIKKRKGKEAKEYKDAGIMPEFVQMSLKPAIGQRYWEEHKDEIYSLDQINLAS---GRTV 270
Query 60 RPPKYFDRIFDVE----------------------YPDESAA-----MKEARKRIAMDAR 92
+PP+YFD++ D E P+E+ + +K+ R++ +
Sbjct 271 KPPRYFDKLEDQEMLMDEIGQIESLKQKFEPDCEMMPEEAESDFMRDIKKKRRKTVLAKT 330
Query 93 QAKLAQTNLSYIDLLAVEERALKERIKSLKRR 124
+A+ TNL + ++ER + + K K R
Sbjct 331 EARWRATNLDLREYYEMQERNFENKNKLAKNR 362
>gi|19424731|ref|NP_598335.1| hypothetical protein Sp-4p1 [Spiroplasma phage 4]
gi|137995|sp|P11334.1|REP_SPV4 RecName: Full=Replication-associated protein ORF2; AltName: Full=Rep
[Spiroplasma phage 4]
Length=320
Score = 56.2 bits (134), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 35/105 (33%), Positives = 59/105 (56%), Gaps = 3/105 (3%)
Query 24 QMSLKPGIGRNYY-DNHPDLYKYEYINVSTAKGGRKFRPPKYFDRIFDVEYPDESAA--M 80
++ + GIG Y+ +N +YK + + +ST KG ++F+ PKYFDR + E+ DE +
Sbjct 211 KLRMSKGIGLKYFMENKERIYKEDSVLISTDKGIKRFKVPKYFDRRMEREWQDEFYLDYI 270
Query 81 KEARKRIAMDARQAKLAQTNLSYIDLLAVEERALKERIKSLKRRL 125
KE R+++A + ++ SY D L E++ L +K L R L
Sbjct 271 KEKREKVAKRTLFQRQIVSSRSYTDYLGDEQKKLNNIVKRLTRPL 315
>gi|575094436|emb|CDL65809.1| unnamed protein product [uncultured bacterium]
Length=340
Score = 53.5 bits (127), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 33/126 (26%), Positives = 66/126 (52%), Gaps = 5/126 (4%)
Query 1 VTKKLTGPAACFYDDHNIEPPFTQMSLKPGIGRNYYDNHPD-LYKYEYINVSTAKGGRKF 59
VTKK+ G Y + + P+ MS KPGIG + + + D L++ +YI ++ G+
Sbjct 219 VTKKMYGNDTKEYQNLGLTAPYACMSRKPGIGMPWLEQNLDKLWEQDYIQLA----GKTA 274
Query 60 RPPKYFDRIFDVEYPDESAAMKEARKRIAMDARQAKLAQTNLSYIDLLAVEERALKERIK 119
P+ FD++ + P+ K+AR++ A++ ++QT+ + ++ ++R L +
Sbjct 275 PIPRAFDKMLEATDPERLWKKKQARQKSAINGALQAMSQTDQTLLEQYETKDRVLMKSFA 334
Query 120 SLKRRL 125
K +L
Sbjct 335 KSKGKL 340
>gi|313766924|gb|ADR80651.1| putative replication initiation protein [Uncultured Microviridae]
Length=285
Score = 47.8 bits (112), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 34/109 (31%), Positives = 55/109 (50%), Gaps = 10/109 (9%)
Query 18 IEPPFTQMSLKPGIGRNYYDNHP-DLYKYEYINVSTAKGGRKFRPPKYFDRIFDVEYPDE 76
+ P + MS KPGIG+ +Y+ + D+Y + GG K P+++D++ + E P++
Sbjct 186 VRPEYATMSRKPGIGKEWYEKYKKDMYPSN--QTPSVGGGVKNGIPRFYDKLMEKEDPEQ 243
Query 77 SAAMKEARKRIAMDARQAKLAQTNLSYIDLLAVEERALKERIKSLKRRL 125
+KE RK A+D T +D V+ A IK+LKR L
Sbjct 244 LEIVKEKRKEFALDNMH---DNTGPRLLDKATVKLAA----IKTLKRDL 285
Lambda K H a alpha
0.319 0.136 0.397 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 429173526870