bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-29_CDS_annotation_glimmer3.pl_2_2
Length=335
Score E
Sequences producing significant alignments: (Bits) Value
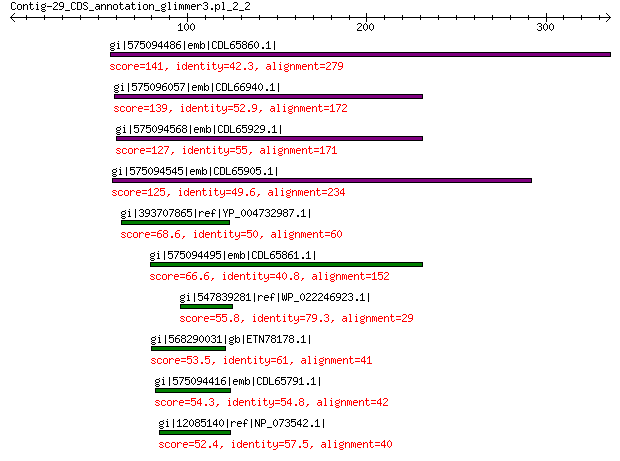
gi|575094486|emb|CDL65860.1| unnamed protein product 141 3e-35
gi|575096057|emb|CDL66940.1| unnamed protein product 139 3e-35
gi|575094568|emb|CDL65929.1| unnamed protein product 127 2e-30
gi|575094545|emb|CDL65905.1| unnamed protein product 125 2e-29
gi|393707865|ref|YP_004732987.1| structural protein VP2 68.6 4e-10
gi|575094495|emb|CDL65861.1| unnamed protein product 66.6 2e-09
gi|547839281|ref|WP_022246923.1| putative minor capsid protein 55.8 8e-06
gi|568290031|gb|ETN78178.1| hypothetical protein NECAME_18237 53.5 1e-05
gi|575094416|emb|CDL65791.1| unnamed protein product 54.3 4e-05
gi|12085140|ref|NP_073542.1| minor capsid protein 52.4 8e-05
>gi|575094486|emb|CDL65860.1| unnamed protein product [uncultured bacterium]
Length=344
Score = 141 bits (356), Expect = 3e-35, Method: Compositional matrix adjust.
Identities = 118/280 (42%), Positives = 168/280 (60%), Gaps = 30/280 (11%)
Query 57 TSANNAWSAQQAEELRKWQEAQTDKAMKYNSQEAEKNRSWQEYMSSTAHQREIADLKAAG 116
T++NNAWSA QA++ +Q +Q ++N EAE +R WQE MS+TAHQREI DL+AAG
Sbjct 72 TASNNAWSAAQAQKQMDFQASQGALVRQFNHDEAELSRLWQERMSNTAHQREIKDLQAAG 131
Query 117 LNPVLSAMggngasvtsgatass-sapsgamgstDTSGSSALVNLLGAMLTSTTELSKMS 175
LNPVLSAMGG+GA VTSG+TAS S PSG+ G TDTS + ALV+LLG+ + + ++ +
Sbjct 132 LNPVLSAMGGSGAPVTSGSTASGYSPPSGSKGDTDTSLAGALVSLLGSSMMAQASMANTA 191
Query 176 TSALTNLAVADKYNSVNKYLGELSSATQLKGYQISAQTALSTANISAAAQRYVSDNNLKG 235
SA T +VADKY +++K + E I +T LS + ISA A RY +D +
Sbjct 192 MSARTQESVADKYTAMSKLVAE-----------IQQETTLSASTISAMASRYAADRSADA 240
Query 236 SLanaaatkiaatihaeaSKYAADKGYLSSENVANINASVNKQLKEMGIKADFDFAQMYP 295
S K+AA+IHA A +Y D ++ ++A+ NA VNK L +MG + DFD + YP
Sbjct 241 S-------KVAASIHAAAQRYGYDVQAMTQRDIASFNAQVNKDLAQMGYQHDFDIKEAYP 293
Query 296 NNLYQMTGATVNNLKGILGDLMSQSAIDSVSSSKGLIKPW 335
+++ G++ L +S + + GL W
Sbjct 294 -----------SSMAGLMASLFGESILGNDKGLSGLSDLW 322
>gi|575096057|emb|CDL66940.1| unnamed protein product [uncultured bacterium]
Length=275
Score = 139 bits (351), Expect = 3e-35, Method: Compositional matrix adjust.
Identities = 91/172 (53%), Positives = 118/172 (69%), Gaps = 11/172 (6%)
Query 59 ANNAWSAQQAEELRKWQEAQTDKAMKYNSQEAEKNRSWQEYMSSTAHQREIADLKAAGLN 118
AN+AW+A+QAE R WQEAQ KAM++NS EA KNR WQE MS+TAHQRE+ DL AAGLN
Sbjct 34 ANSAWNAEQAEIQRDWQEAQNAKAMQFNSMEAAKNRKWQEMMSNTAHQREVKDLMAAGLN 93
Query 119 PVLSAMggngasvtsgatasssapsgamgstDTSGSSALVNLLGAMLTSTTELSKMSTSA 178
PVLSAM GNGA+V SGATAS +GA G DTS S A+ NLLG++L+++T + + +A
Sbjct 94 PVLSAMNGNGAAVGSGATASGVTSAGAKGEADTSTSGAIANLLGSILSASTAIQAANVNA 153
Query 179 LTNLAVADKYNSVNKYLGELSSATQLKGYQISAQTALSTANISAAAQRYVSD 230
T AVADKY ++++ + E++ A L +A I A A RY +D
Sbjct 154 RTQEAVADKYTAMSQIVAEINKA-----------ATLGSAGIHAGATRYAAD 194
>gi|575094568|emb|CDL65929.1| unnamed protein product [uncultured bacterium]
Length=310
Score = 127 bits (319), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 94/182 (52%), Positives = 121/182 (66%), Gaps = 11/182 (6%)
Query 60 NNAWSAQQAEELRKWQEAQTDKAMKYNSQEAEKNRSWQEYMSSTAHQREIADLKAAGLNP 119
N A S Q+AE LR WQE Q AM++N+ EAEKNR+WQE MS+TAHQRE+ DL AAGLNP
Sbjct 53 NTARSVQEAESLRTWQEEQNRIAMQFNAAEAEKNRNWQEIMSNTAHQREVNDLMAAGLNP 112
Query 120 VLSAMggngasvtsgatasssapsgamgstDTSGSSALVNLLGAMLTSTTELSKMSTSAL 179
VLSA GGNGA+VTSGATAS SGA G DTS SSA+V +LG+ML+S T ++ +TSA+
Sbjct 113 VLSAGGGNGAAVTSGATASGVTSSGAKGDVDTSASSAVVGILGSMLSSLTNIANANTSAI 172
Query 180 TNLAVADKYNSVNKYLGELSSATQLK-----GYQISAQTA------LSTANISAAAQRYV 228
T++A +K +N+ + ++ LK G AQ A L+ A + A A RY
Sbjct 173 TSMANTEKLGQINQLIAHANNENALKVAETYGKYGVAQAATAGRYSLNAAQVHADATRYS 232
Query 229 SD 230
+D
Sbjct 233 AD 234
>gi|575094545|emb|CDL65905.1| unnamed protein product [uncultured bacterium]
Length=325
Score = 125 bits (313), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 116/256 (45%), Positives = 157/256 (61%), Gaps = 40/256 (16%)
Query 58 SANNAWSAQQAEELRKWQEAQTDKAMKYNSQEAEKNRSWQEYMSSTAHQREIADLKAAGL 117
S N+A++A QA R WQ+ Q + AM+++S EA KNR WQ YMS+TAHQRE+ADLKAAGL
Sbjct 32 SDNSAFNASQAAANRNWQQQQNNIAMQFSSAEAAKNRDWQSYMSNTAHQREVADLKAAGL 91
Query 118 NPVLSAMggngasvtsgatasssapsgamgstDTSGSSALVNLLGAMLTSTTELSKMSTS 177
NPVLSAMGGNGA+VTSGATA SG S DTS ++ALV LLG++L + T ++ +T+
Sbjct 92 NPVLSAMGGNGAAVTSGATAQGYTSSGGQASADTSATAALVGLLGSLLNAQTSIANTATN 151
Query 178 ALTNLAVADKYNSVNKYLGELSSATQLKGYQISAQTALSTANISAAAQRYVSDNNLKGSL 237
A+ NL+VADKY S +Y ++ GY A T+ S AN++A A R+ S+N L S
Sbjct 152 AVANLSVADKYTSATRYAADV-------GY---AGTSYS-ANVAAYASRFASNNALAAS- 199
Query 238 anaaatkiaatihaeaSKYAADKGYLSSE----------------------NVANINASV 275
K A+ ASKYA+D+ YL+S+ ++A NA+V
Sbjct 200 ------KYASDNSRAASKYASDQSYLASKFASILQSNTAKYNIDTRTATDRDLAEFNAAV 253
Query 276 NKQLKEMGIKADFDFA 291
N+ L++ I A F A
Sbjct 254 NRDLQKNEIDAKFSLA 269
>gi|393707865|ref|YP_004732987.1| structural protein VP2 [Microviridae phi-CA82]
gi|311336637|gb|ADP89808.1| structural protein VP2 [Microviridae phi-CA82]
Length=234
Score = 68.6 bits (166), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 30/60 (50%), Positives = 44/60 (73%), Gaps = 0/60 (0%)
Query 63 WSAQQAEELRKWQEAQTDKAMKYNSQEAEKNRSWQEYMSSTAHQREIADLKAAGLNPVLS 122
W QA++ KW QT+K+ ++N+QEA+KNR WQE MS+TA QR++ D + AGLNP+ +
Sbjct 7 WMTAQADKQNKWNAEQTEKSNQFNAQEAQKNRDWQEQMSNTALQRKMQDAEKAGLNPIFA 66
>gi|575094495|emb|CDL65861.1| unnamed protein product [uncultured bacterium]
Length=266
Score = 66.6 bits (161), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 62/152 (41%), Positives = 84/152 (55%), Gaps = 14/152 (9%)
Query 79 TDKAMKYNSQEAEKNRSWQEYMSSTAHQREIADLKAAGLNPVLSAMggngasvtsgatas 138
TDK + YN+Q A + ++QE MSSTAHQRE+ DL AAGLNPVLSA +
Sbjct 60 TDKLLNYNTQSAREQMAFQERMSSTAHQREVKDLIAAGLNPVLSA-----------GGSG 108
Query 139 ssapsgamgstDTSGSSALVNLLGAMLTSTTELSKMSTSALTNLAVADKYNSVNKYLGEL 198
+SAPSGAM + D+S SA N A L +++ + N A D ++NKY ++
Sbjct 109 ASAPSGAMATADSSMMSAKAN---AALQKRIVNAQLKNAKDINKAQLDAQKAMNKYSVDV 165
Query 199 SSATQLKGYQISAQTALSTANISAAAQRYVSD 230
+ T L QISA + A +AAA Y S+
Sbjct 166 GAQTSLANAQISASASKFGAMQAAAASMYGSN 197
>gi|547839281|ref|WP_022246923.1| putative minor capsid protein [Clostridium sp. CAG:306]
gi|524476581|emb|CDC18646.1| putative minor capsid protein [Clostridium sp. CAG:306]
Length=236
Score = 55.8 bits (133), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 23/29 (79%), Positives = 27/29 (93%), Gaps = 0/29 (0%)
Query 96 WQEYMSSTAHQREIADLKAAGLNPVLSAM 124
+QE MSSTAHQRE+ DL+AAGLNP+LSAM
Sbjct 41 FQERMSSTAHQREVKDLRAAGLNPILSAM 69
>gi|568290031|gb|ETN78178.1| hypothetical protein NECAME_18237 [Necator americanus]
Length=112
Score = 53.5 bits (127), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 25/41 (61%), Positives = 30/41 (73%), Gaps = 0/41 (0%)
Query 80 DKAMKYNSQEAEKNRSWQEYMSSTAHQREIADLKAAGLNPV 120
D A K N + + +WQE MS+TAHQRE ADLKAAGLNP+
Sbjct 28 DAANKANRKMMREQMAWQERMSNTAHQREQADLKAAGLNPI 68
>gi|575094416|emb|CDL65791.1| unnamed protein product [uncultured bacterium]
Length=311
Score = 54.3 bits (129), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 23/42 (55%), Positives = 31/42 (74%), Gaps = 0/42 (0%)
Query 82 AMKYNSQEAEKNRSWQEYMSSTAHQREIADLKAAGLNPVLSA 123
A +YN +EA+ RSW + M TA+Q + DLKAAGLNP+L+A
Sbjct 144 AQRYNREEAQAERSWAQSMRQTAYQDTVKDLKAAGLNPILAA 185
>gi|12085140|ref|NP_073542.1| minor capsid protein [Bdellovibrio phage phiMH2K]
gi|75089169|sp|Q9G055.1|H_BPPHM RecName: Full=Minor spike protein H; AltName: Full=H protein;
AltName: Full=Pilot protein; AltName: Full=Protein VP2; Short=VP2
[Bdellovibrio phage phiMH2K]
gi|12017988|gb|AAG45344.1|AF306496_5 Vp2 [Bdellovibrio phage phiMH2K]
Length=199
Score = 52.4 bits (124), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 23/40 (58%), Positives = 30/40 (75%), Gaps = 0/40 (0%)
Query 84 KYNSQEAEKNRSWQEYMSSTAHQREIADLKAAGLNPVLSA 123
+ N EA +NR WQE MS++AHQRE DL+ AGLN +L+A
Sbjct 40 RENQAEAARNRKWQEQMSNSAHQREANDLQTAGLNRLLTA 79
Lambda K H a alpha
0.306 0.119 0.324 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1927902993225