bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-30_CDS_annotation_glimmer3.pl_2_2
Length=278
Score E
Sequences producing significant alignments: (Bits) Value
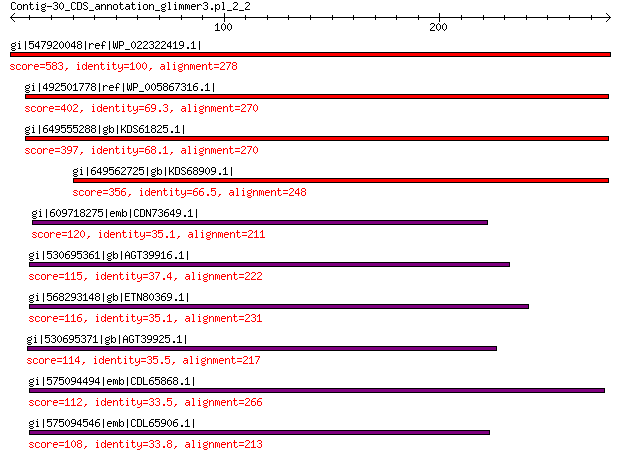
gi|547920048|ref|WP_022322419.1| putative replication protein 583 0.0
gi|492501778|ref|WP_005867316.1| hypothetical protein 402 6e-138
gi|649555288|gb|KDS61825.1| hypothetical protein M095_3808 397 8e-136
gi|649562725|gb|KDS68909.1| hypothetical protein M096_3339 356 2e-120
gi|609718275|emb|CDN73649.1| conserved hypothetical protein 120 1e-28
gi|530695361|gb|AGT39916.1| replication initiator 115 9e-27
gi|568293148|gb|ETN80369.1| hypothetical protein NECAME_18023 116 1e-26
gi|530695371|gb|AGT39925.1| replication initiator 114 5e-26
gi|575094494|emb|CDL65868.1| unnamed protein product 112 2e-25
gi|575094546|emb|CDL65906.1| unnamed protein product 108 7e-24
>gi|547920048|ref|WP_022322419.1| putative replication protein [Parabacteroides merdae CAG:48]
gi|524592960|emb|CDD13572.1| putative replication protein [Parabacteroides merdae CAG:48]
Length=278
Score = 583 bits (1504), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 278/278 (100%), Positives = 278/278 (100%), Gaps = 0/278 (0%)
Query 1 MYLASVEAKVPCGWCVNCRQNKRQSWVYRLQAEAKEYPLSLFVTLTYDDEHLPIERIGSD 60
MYLASVEAKVPCGWCVNCRQNKRQSWVYRLQAEAKEYPLSLFVTLTYDDEHLPIERIGSD
Sbjct 1 MYLASVEAKVPCGWCVNCRQNKRQSWVYRLQAEAKEYPLSLFVTLTYDDEHLPIERIGSD 60
Query 61 LFQTNVAVVSKRDVQLFMKRLRKKYEDYKMRYFVTSEYGAKNGRPHYHMILFGFPFTGKM 120
LFQTNVAVVSKRDVQLFMKRLRKKYEDYKMRYFVTSEYGAKNGRPHYHMILFGFPFTGKM
Sbjct 61 LFQTNVAVVSKRDVQLFMKRLRKKYEDYKMRYFVTSEYGAKNGRPHYHMILFGFPFTGKM 120
Query 121 AGDLLAECWQNGFVQAHPLTIKEIAYVCKYMYEKSMCPEILRDEKKYKPFMLCSRNPGIG 180
AGDLLAECWQNGFVQAHPLTIKEIAYVCKYMYEKSMCPEILRDEKKYKPFMLCSRNPGIG
Sbjct 121 AGDLLAECWQNGFVQAHPLTIKEIAYVCKYMYEKSMCPEILRDEKKYKPFMLCSRNPGIG 180
Query 181 FGFMKADIIEFYRRHPRDYVRAWAGHKMAMPRYYADKLYDDDMKAFLKEMREEFFRHKMF 240
FGFMKADIIEFYRRHPRDYVRAWAGHKMAMPRYYADKLYDDDMKAFLKEMREEFFRHKMF
Sbjct 181 FGFMKADIIEFYRRHPRDYVRAWAGHKMAMPRYYADKLYDDDMKAFLKEMREEFFRHKMF 240
Query 241 NEWIDYCARENPILTDLMQLEQREEYEKRMNERLRCKM 278
NEWIDYCARENPILTDLMQLEQREEYEKRMNERLRCKM
Sbjct 241 NEWIDYCARENPILTDLMQLEQREEYEKRMNERLRCKM 278
>gi|492501778|ref|WP_005867316.1| hypothetical protein [Parabacteroides distasonis]
gi|409230407|gb|EKN23271.1| hypothetical protein HMPREF1059_03256 [Parabacteroides distasonis
CL09T03C24]
Length=284
Score = 402 bits (1034), Expect = 6e-138, Method: Compositional matrix adjust.
Identities = 187/272 (69%), Positives = 223/272 (82%), Gaps = 2/272 (1%)
Query 8 AKVPCGWCVNCRQNKRQSWVYRLQAEAKEYPLSLFVTLTYDDEHLPIERIGSDLFQTNVA 67
VPCG CVNCR+NKRQSWVYRLQAEA EYP SLFVTLTYDDEH+P IG DLF++ V
Sbjct 13 GAVPCGRCVNCRKNKRQSWVYRLQAEADEYPFSLFVTLTYDDEHMPTAMIGEDLFKSTVG 72
Query 68 VVSKRDVQLFMKRLRKKYEDYKMRYFVTSEYGAKNGRPHYHMILFGFPFTGKMAGDLLAE 127
VVSKRD+QLFMKRLRKKY+ Y++RYF+TSEYG++ GRPHYHMILFGFPFTGK GDLLAE
Sbjct 73 VVSKRDIQLFMKRLRKKYDQYRLRYFLTSEYGSQGGRPHYHMILFGFPFTGKHGGDLLAE 132
Query 128 CWQNGFVQAHPLTIKEIAYVCKYMYEKSMCPEILRDEKKYKPFMLCSRNPGIGFGFMKAD 187
CW+NGFVQAHPLT KEIAYV KYMYEKSM P+IL+D K+Y+PFMLCSR PGIG+ F++
Sbjct 133 CWKNGFVQAHPLTTKEIAYVTKYMYEKSMVPDILKDVKEYQPFMLCSRIPGIGYHFLREQ 192
Query 188 IIEFYRRHPRDYVRAWAGHKMAMPRYYADKLYDDDMKAFLKEMREEFFRHKMFNEWIDYC 247
I++FYR HPRDYVRA+ G +MAMPRYYADKLYDDDMK +LKE+RE FF ++M EW Y
Sbjct 193 ILDFYRLHPRDYVRAFNGMRMAMPRYYADKLYDDDMKEYLKELREAFFINQMQQEWHHYI 252
Query 248 ARENPI--LTDLMQLEQREEYEKRMNERLRCK 277
+ + D ++ E + +YE+R E+L+ K
Sbjct 253 NTSPRLRYIADQLETESKLDYERRAEEKLKLK 284
>gi|649555288|gb|KDS61825.1| hypothetical protein M095_3808 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649560564|gb|KDS66872.1| hypothetical protein M095_2449 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649561011|gb|KDS67298.1| hypothetical protein M095_2409 [Parabacteroides distasonis str.
3999B T(B) 4]
Length=284
Score = 397 bits (1020), Expect = 8e-136, Method: Compositional matrix adjust.
Identities = 184/272 (68%), Positives = 220/272 (81%), Gaps = 2/272 (1%)
Query 8 AKVPCGWCVNCRQNKRQSWVYRLQAEAKEYPLSLFVTLTYDDEHLPIERIGSDLFQTNVA 67
VPCG CVNCR+NKRQSWVYRLQAEA EYP SLFVTLTYDDEH+P IG DLF+T V
Sbjct 13 GAVPCGRCVNCRKNKRQSWVYRLQAEADEYPFSLFVTLTYDDEHIPTAMIGEDLFKTTVG 72
Query 68 VVSKRDVQLFMKRLRKKYEDYKMRYFVTSEYGAKNGRPHYHMILFGFPFTGKMAGDLLAE 127
VVSKRD+QLFMKRLRKKY Y++RYF+TSEYG++ GRPHYHMILFGFPFTGK GDLLAE
Sbjct 73 VVSKRDIQLFMKRLRKKYAQYRLRYFLTSEYGSQGGRPHYHMILFGFPFTGKHGGDLLAE 132
Query 128 CWQNGFVQAHPLTIKEIAYVCKYMYEKSMCPEILRDEKKYKPFMLCSRNPGIGFGFMKAD 187
CW+NGFVQAHPLT KEI+YV KYMYEKSM P+IL+ K+Y+PFMLCS+ PGIG+ F++
Sbjct 133 CWKNGFVQAHPLTTKEISYVTKYMYEKSMIPDILKGVKEYQPFMLCSKMPGIGYHFLREQ 192
Query 188 IIEFYRRHPRDYVRAWAGHKMAMPRYYADKLYDDDMKAFLKEMREEFFRHKMFNEWIDYC 247
I++FYR HPRDYVRA+ G +MAMPRYYADKLYDDDMK +LKE+RE FF ++M EW Y
Sbjct 193 ILDFYRLHPRDYVRAFNGMRMAMPRYYADKLYDDDMKEYLKELREAFFINQMQQEWYHYI 252
Query 248 ARENPI--LTDLMQLEQREEYEKRMNERLRCK 277
+ + D ++ E + YE+R ++L+ K
Sbjct 253 NTSPRLRYIADQLETESKLAYERRAEDKLKLK 284
>gi|649562725|gb|KDS68909.1| hypothetical protein M096_3339 [Parabacteroides distasonis str.
3999B T(B) 6]
Length=250
Score = 356 bits (914), Expect = 2e-120, Method: Compositional matrix adjust.
Identities = 165/250 (66%), Positives = 201/250 (80%), Gaps = 2/250 (1%)
Query 30 LQAEAKEYPLSLFVTLTYDDEHLPIERIGSDLFQTNVAVVSKRDVQLFMKRLRKKYEDYK 89
+QAEA EYP SLFVTLTYDDEH+P IG DLF+T V VVSKRD+QLFMKRLRKKY Y+
Sbjct 1 MQAEADEYPFSLFVTLTYDDEHIPTAMIGEDLFKTTVGVVSKRDIQLFMKRLRKKYAQYR 60
Query 90 MRYFVTSEYGAKNGRPHYHMILFGFPFTGKMAGDLLAECWQNGFVQAHPLTIKEIAYVCK 149
+RYF+TSEYG++ GRPHYHMILFGFPFTGK GDLLAECW+NGFVQAHPLT KEI+YV K
Sbjct 61 LRYFLTSEYGSQGGRPHYHMILFGFPFTGKHGGDLLAECWKNGFVQAHPLTTKEISYVTK 120
Query 150 YMYEKSMCPEILRDEKKYKPFMLCSRNPGIGFGFMKADIIEFYRRHPRDYVRAWAGHKMA 209
YMYEKSM P+IL+ K+Y+PFMLCS+ PGIG+ F++ I++FYR HPRDYVRA+ G +MA
Sbjct 121 YMYEKSMIPDILKGVKEYQPFMLCSKMPGIGYHFLREQILDFYRLHPRDYVRAFNGMRMA 180
Query 210 MPRYYADKLYDDDMKAFLKEMREEFFRHKMFNEWIDYCARENPI--LTDLMQLEQREEYE 267
MPRYYADKLYDDDMK +LKE+RE FF ++M EW Y + + D ++ E + YE
Sbjct 181 MPRYYADKLYDDDMKEYLKELREAFFINQMQQEWYHYINTSPRLRYIADQLETESKLAYE 240
Query 268 KRMNERLRCK 277
+R ++L+ K
Sbjct 241 RRAEDKLKLK 250
>gi|609718275|emb|CDN73649.1| conserved hypothetical protein [Elizabethkingia anophelis]
Length=265
Score = 120 bits (300), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 74/211 (35%), Positives = 104/211 (49%), Gaps = 14/211 (7%)
Query 11 PCGWCVNCRQNKRQSWVYRLQAEAKEYPLSLFVTLTYDDEHLPIERIGSDLFQTNVAVVS 70
PCG C+ CR+ + SW RL E K + FVTLTY D +LP G + +
Sbjct 24 PCGKCLECRKARTNSWFARLTEELKVSKSAHFVTLTYSDVYLPYSDNG-------LISLD 76
Query 71 KRDVQLFMKRLRKKYEDYKMRYFVTSEYGAKNGRPHYHMILFGFPFTGKMAGDLLAECWQ 130
RD QLFMKR R K + K++YF+ EYGA+ RPHYH I+FG G+ W+
Sbjct 77 YRDFQLFMKRAR-KLQKSKIKYFLVGEYGAQTYRPHYHAIVFGVENIDAFLGE-----WR 130
Query 131 NGFVQAHPLTIKEIAYVCKYMYEKSMCPEILRDEKKYKPFMLCSRNPGIGFGFMKADIIE 190
G V A +T K I Y KY KS+ +D + + G+G + +I+
Sbjct 131 MGNVHAGTVTAKSIYYTLKYC-TKSITEGPDKDPDDDRKPEKALMSKGLGLSHLTESMIK 189
Query 191 FYRRHPRDYVRAWAGHKMAMPRYYADKLYDD 221
+Y+ G +A+PRYY DK++ D
Sbjct 190 YYKDDVSRSFSLLGGTTIALPRYYRDKVFSD 220
>gi|530695361|gb|AGT39916.1| replication initiator [Marine gokushovirus]
Length=289
Score = 115 bits (289), Expect = 9e-27, Method: Compositional matrix adjust.
Identities = 83/247 (34%), Positives = 122/247 (49%), Gaps = 40/247 (16%)
Query 10 VPCGWCVNCRQNKRQSWVYRLQAEAKEYPLSLFVTLTYDDEHLPIERIGSDLFQTNVAVV 69
+PCG C+ CR + + W R EA+ + + F+TLT+D+EH+ + N +
Sbjct 30 LPCGQCIGCRLDYSRQWAIRCVHEAQTHEDNCFITLTFDNEHIAKRK--------NPESL 81
Query 70 SKRDVQLFMKRLRKKYEDYKMRYFVTSEYGAKNGRPHYHMILFGFPFTGKM----AGDL- 124
+ Q FMKRLRKKY +K+R+F EYG +N RPHYH +LFG F K GD
Sbjct 82 DNTEFQRFMKRLRKKYP-HKIRFFHCGEYGDQNKRPHYHALLFGHDFKDKKLWSNKGDFK 140
Query 125 ------LAECWQNGFVQAHPLTIKEIAYVCKYMYEKSM--------------CPEILRDE 164
LAE W GF ++ AY +Y+ +K E++ +
Sbjct 141 LFVSQELAELWPYGFHTIGAVSFDTAAYCARYVMKKVTGDAAASHYREVDLETGEVINEI 200
Query 165 KKYKPFMLCSRNPGIGFGFMKADIIEFYRRHPRDYVRAWAGHKMAMPRYYADKLYDDDMK 224
K + SR PGIG+ + + ++ H DY+ G+K+ PRYY DKL D++M
Sbjct 201 K--PEYCTMSRMPGIGYEWYQK--YGYHDCHKHDYI-VINGYKVRPPRYY-DKLCDEEMF 254
Query 225 AFLKEMR 231
A +KE R
Sbjct 255 AQIKETR 261
>gi|568293148|gb|ETN80369.1| hypothetical protein NECAME_18023 [Necator americanus]
Length=345
Score = 116 bits (291), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 81/238 (34%), Positives = 126/238 (53%), Gaps = 22/238 (9%)
Query 10 VPCGWCVNCRQNKRQSWVYRLQAEAKEYPLSLFVTLTYDDEHLPIERIGSDLFQTNVAVV 69
VPCG C C++ + SWV+RL E ++ + FVTLTYD +PI + G F T +
Sbjct 20 VPCGRCPPCKRRRVDSWVFRLLQEELQHENASFVTLTYDTRFVPISKNG---FMT----L 72
Query 70 SKRDVQLFMKRLRKKYEDYKMRYFVTSEYGAKNGRPHYHMILFGFPFTGKMAGDLLAECW 129
+ + +MKRLRK K++Y++ EYG++ RPHYH I+FG P L A+ W
Sbjct 73 DRGEFPRYMKRLRKLVPGRKLKYYMCGEYGSQRFRPHYHAIIFGVP-----QDSLFADAW 127
Query 130 Q-NG----FVQAHPLTIKEIAYVCKYMYEKSMCPEILRDEKKYKPFMLCSRNPGIGFGFM 184
NG V +T K IAY KY+ + + + RD+ + F L S+ G+G ++
Sbjct 128 TLNGDSLGGVVVGTVTGKSIAYTMKYIDKSTWKQKHGRDD-RVPEFSLMSK--GMGVSYL 184
Query 185 KADIIEFYRRH-PRDYVRAWAGHKMAMPRYYADKLY-DDDMKAFLKEMREEFFRHKMF 240
++E+++ R + G ++AMPRYY K+Y DDD+K + + E R +
Sbjct 185 TPQMVEYHKEDISRLFCTREGGSRIAMPRYYRQKIYSDDDLKKQVVLIAESVERQEQL 242
>gi|530695371|gb|AGT39925.1| replication initiator [Marine gokushovirus]
Length=316
Score = 114 bits (285), Expect = 5e-26, Method: Compositional matrix adjust.
Identities = 77/240 (32%), Positives = 114/240 (48%), Gaps = 38/240 (16%)
Query 9 KVPCGWCVNCRQNKRQSWVYRLQAEAKEYPLSLFVTLTYDDEHLPIERIGSDLFQTNVAV 68
++PC C+ CR K + W R EAK Y + F+TLTY+ +HLP L ++
Sbjct 48 QIPCNQCIGCRLEKSRQWALRCTHEAKLYKNNSFITLTYNSDHLP-------LTNNSLPT 100
Query 69 VSKRDVQLFMKRLRKKYEDYKMRYFVTSEYGAKNGRPHYHMILFGFPFTGKM-------- 120
++ R QLF+KRLRKKY + +R++ EYG N RPHYH +LF F K
Sbjct 101 LNLRHFQLFLKRLRKKYSNKTIRFYHCGEYGDMNHRPHYHALLFNHDFEDKKLWKIHKDQ 160
Query 121 ---AGDLLAECWQN-------GFVQAHPLTIKEIAYVCKYMYEKSMCPEILRD--EKKYK 168
++L W + GF LT AYV +Y +K + + D + +
Sbjct 161 NYYTSEVLDGLWTDPKTKSNMGFSTIGDLTFDSAAYVARYCLKK-ITGKNAEDYYQGRVP 219
Query 169 PFMLCSRNPGIGFGFM---KADIIEFYRRHPRDYVRAWAGHKMAMPRYYADKLYDDDMKA 225
+ SR PGIG G++ K+D+ +P ++ G KM P+YY + D KA
Sbjct 220 EYATMSRRPGIGNGWLDKFKSDV------YPSGFI-IHEGQKMQPPKYYDRVTNETDEKA 272
>gi|575094494|emb|CDL65868.1| unnamed protein product [uncultured bacterium]
Length=348
Score = 112 bits (281), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 89/290 (31%), Positives = 137/290 (47%), Gaps = 34/290 (12%)
Query 10 VPCGWCVNCRQNKRQSWVYRLQAEAKEYPLSLFVTLTYDDEHLPI-ERIGSDLFQTNV-A 67
+PCG CV CR + W R E+ + S F+TLTYDD++LP+ E I D + N A
Sbjct 62 IPCGKCVGCRLAYSRQWADRCMLESSYHTHSYFLTLTYDDDNLPLSESINQDTGEINYNA 121
Query 68 VVSKRDVQLFMKRLRKKYE-----DYKMRYFVTSEYGAKNGRPHYHMILFGFPFTG---- 118
+ K+D+Q F+KRLR+ E + ++YF EYG++ RPHYHMIL+GFP
Sbjct 122 TLVKKDIQDFIKRLRRFCEYNIDDNLHIKYFCAGEYGSQTFRPHYHMILYGFPINDLKLY 181
Query 119 KMAGD--------LLAECWQNGFVQAHPLTIKEIAYVCKYMYEK--SMCPEILRDEKKYK 168
KM+ D + + W+ GFV +T AY +Y+ +K +I +D
Sbjct 182 KMSLDGYNYYNSATIDKLWKKGFVVIGEVTWDTCAYTARYILKKQYGSGAQIYKDYNILP 241
Query 169 PFMLCSRNPGIGFGFMKADIIEFYRRHPRDYVRAWAGHK---MAMPRYYADKLYDDDMKA 225
F S P I + + + + + DY+ K M P+Y+ +KL + + +
Sbjct 242 EFTCMSTKPAIAREYYEDNKDKIF---DSDYIFLGTKEKSIQMKPPKYF-EKLLEKENED 297
Query 226 FLKEMREEFFRHKMFNEWIDYCARENPILTDLMQLEQREEYEKRMNERLR 275
KE R+ H E C R D + + Q E E +N R++
Sbjct 298 VFKERRD---LHASLAEDFS-CLRNLSTSHDYLGMLQME--EDNLNARIK 341
>gi|575094546|emb|CDL65906.1| unnamed protein product [uncultured bacterium]
Length=351
Score = 108 bits (270), Expect = 7e-24, Method: Compositional matrix adjust.
Identities = 72/243 (30%), Positives = 112/243 (46%), Gaps = 31/243 (13%)
Query 10 VPCGWCVNCRQNKRQSWVYRLQAEAKEYPLSLFVTLTYDDEHLPIERIGSDLFQTNVA-V 68
+PCG C+ CR + + W RL E + + ++FVTLTY + ++P + N +
Sbjct 56 LPCGQCIGCRLDYSRRWADRLMLELQYHTAAIFVTLTYSELNVPKHHYQTPDGDVNTSYS 115
Query 69 VSKRDVQLFMKRLRKKYEDYKMRYFVTSEYGAKNGRPHYHMILFGFPFTG-------KMA 121
+ KRDVQLF KRLRK Y D K+R+F++ EYG K RPHYH I+FG F + A
Sbjct 116 LDKRDVQLFFKRLRKMYPDTKIRFFLSGEYGPKTFRPHYHAIIFGVDFAHDRYVWRVRRA 175
Query 122 GDLLAECWQN--------------------GFVQAHPLTIKEIAYVCKYMYEK--SMCPE 159
++ +++ G V+ ++ AYV +Y+ +K +
Sbjct 176 DNMFVNYYRSPSLERAWSVYNNDVGDYVPIGNVEFSDVSWHTCAYVARYVTKKLTGNLAQ 235
Query 160 ILRDEKKYKPFMLCSRNPGIGFGFMKADIIEFYRRHPRDYVRAWAGHKMAMPRYYADKLY 219
PF L SR PGI + + ++ Y + G M P+Y+ D Y
Sbjct 236 FYTTFNLTPPFSLMSRKPGIAYQYYADHGLDIYDNDKINISTERGGLSMLPPKYF-DHFY 294
Query 220 DDD 222
+ D
Sbjct 295 ELD 297
Lambda K H a alpha
0.326 0.140 0.448 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1373811661332