bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-30_CDS_annotation_glimmer3.pl_2_3
Length=290
Score E
Sequences producing significant alignments: (Bits) Value
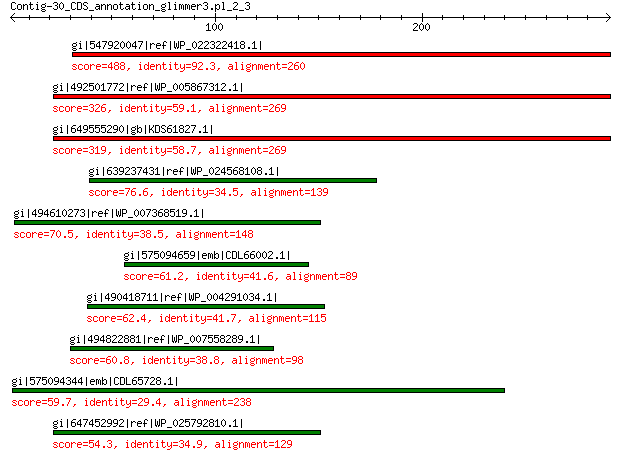
gi|547920047|ref|WP_022322418.1| putative uncharacterized protein 488 7e-172
gi|492501772|ref|WP_005867312.1| hypothetical protein 326 1e-107
gi|649555290|gb|KDS61827.1| hypothetical protein M095_3809 319 4e-105
gi|639237431|ref|WP_024568108.1| hypothetical protein 76.6 5e-13
gi|494610273|ref|WP_007368519.1| hypothetical protein 70.5 2e-10
gi|575094659|emb|CDL66002.1| unnamed protein product 61.2 5e-08
gi|490418711|ref|WP_004291034.1| hypothetical protein 62.4 8e-08
gi|494822881|ref|WP_007558289.1| hypothetical protein 60.8 2e-07
gi|575094344|emb|CDL65728.1| unnamed protein product 59.7 6e-07
gi|647452992|ref|WP_025792810.1| hypothetical protein 54.3 4e-05
>gi|547920047|ref|WP_022322418.1| putative uncharacterized protein [Parabacteroides merdae CAG:48]
gi|524592959|emb|CDD13571.1| putative uncharacterized protein [Parabacteroides merdae CAG:48]
Length=259
Score = 488 bits (1256), Expect = 7e-172, Method: Compositional matrix adjust.
Identities = 240/260 (92%), Positives = 250/260 (96%), Gaps = 1/260 (0%)
Query 31 MQLAKYQNNWQTAENEKAYARSVEMWNMQNQYNSPTAQMSRLRQAGLNPNLVYGSGVTGN 90
MQLAKYQNNWQTAENEKAYARSVEMWNMQNQYNSPTAQMSRLRQAGLNPNLVYGSGVTGN
Sbjct 1 MQLAKYQNNWQTAENEKAYARSVEMWNMQNQYNSPTAQMSRLRQAGLNPNLVYGSGVTGN 60
Query 91 SAGSAPQYQPAKIQRATMEPYRGWNLGLSDAASMYMAMRQNKAQVENMEAQNKLIKEQAR 150
SAGSAPQYQPAKIQRATMEPYRGWNLGLSDAASMYMAMRQNKAQVENMEAQNKLIKEQAR
Sbjct 61 SAGSAPQYQPAKIQRATMEPYRGWNLGLSDAASMYMAMRQNKAQVENMEAQNKLIKEQAR 120
Query 151 TEGIRQGNIAMSTARSGFDLNLARELRNVSIDRAIAEKNLSEASAAGAWTGANQKVLQYE 210
TEGIRQGNIAMSTARSGFDLNLARELRNVSIDRAIAEKNLSEASAAGAWTGANQKVLQYE
Sbjct 121 TEGIRQGNIAMSTARSGFDLNLARELRNVSIDRAIAEKNLSEASAAGAWTGANQKVLQYE 180
Query 211 LDRTLFDNKIKLSNAEYSTAMEALRKLRQDNDINAFRYSMERVFGNSSDVKDVASELVKR 270
LDRTLFDNKIKLSNA+Y+TAME +RKL+QDNDINAFR MER+FG+SSD K+VASEL KR
Sbjct 181 LDRTLFDNKIKLSNAQYATAMEGIRKLQQDNDINAFRNRMERLFGDSSDAKNVASELFKR 240
Query 271 MGLLLMRPDIKELDQMFNPK 290
M + + R D +ELDQMFNPK
Sbjct 241 MMMYMFR-DSRELDQMFNPK 259
>gi|492501772|ref|WP_005867312.1| hypothetical protein [Parabacteroides distasonis]
gi|409230405|gb|EKN23269.1| hypothetical protein HMPREF1059_03254 [Parabacteroides distasonis
CL09T03C24]
Length=288
Score = 326 bits (835), Expect = 1e-107, Method: Compositional matrix adjust.
Identities = 159/269 (59%), Positives = 207/269 (77%), Gaps = 7/269 (3%)
Query 22 AQRQANIQNMQLAKYQNNWQTAENEKAYARSVEMWNMQNQYNSPTAQMSRLRQAGLNPNL 81
A + N NM++AKYQ WQ ENEKAY RS+ MWN+QN+YNSPT QM+R+R AGLNPNL
Sbjct 27 AVQDTNKANMEIAKYQAQWQQQENEKAYQRSLNMWNLQNEYNSPTQQMARIRAAGLNPNL 86
Query 82 VYGSGVTGNSAGSAPQYQPAKIQRATMEPYRGWNLGLSDAASMYMAMRQNKAQVENMEAQ 141
VYG+GVTGNS+GS PQY+PAK TM+ YRGWNLG+SDA S ++A R KAQV+NMEAQ
Sbjct 87 VYGNGVTGNSSGSTPQYEPAKFNAPTMQAYRGWNLGISDAISQFLAYRTVKAQVDNMEAQ 146
Query 142 NKLIKEQARTEGIRQGNIAMSTARSGFDLNLARELRNVSIDRAIAEKNLSEASAAGAWTG 201
N LI++Q TE +Q NIA ST+RS FDLN+A+EL++VS+ AIA+ N +A AA WT
Sbjct 147 NSLIRQQTATEATKQANIAASTSRSEFDLNMAKELKDVSVSSAIADMNQKQAGAAQGWTK 206
Query 202 ANQKVLQYELDRTLFDNKIKLSNAEYSTAMEALRKLRQDNDINAFRYSMERVFGNSSDVK 261
AN++V+QYELD+ LFDNKIKLSN EY ++++R+L+QDNDINAFR MERV G SS
Sbjct 207 ANREVIQYELDKALFDNKIKLSNQEYLRVLQSVRQLQQDNDINAFRNEMERVTGKSS--- 263
Query 262 DVASELVKRMGLLLMRPDIKELDQMFNPK 290
A+++++R+ ++ +RP D++FNPK
Sbjct 264 -FATDMLRRL-IMALRPSAN--DRLFNPK 288
>gi|649555290|gb|KDS61827.1| hypothetical protein M095_3809 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649557306|gb|KDS63785.1| hypothetical protein M095_3404 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649559158|gb|KDS65545.1| hypothetical protein M096_4689 [Parabacteroides distasonis str.
3999B T(B) 6]
gi|649560567|gb|KDS66875.1| hypothetical protein M095_2448 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649561016|gb|KDS67303.1| hypothetical protein M095_2410 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649562727|gb|KDS68911.1| hypothetical protein M096_3341 [Parabacteroides distasonis str.
3999B T(B) 6]
Length=288
Score = 319 bits (818), Expect = 4e-105, Method: Compositional matrix adjust.
Identities = 158/269 (59%), Positives = 204/269 (76%), Gaps = 7/269 (3%)
Query 22 AQRQANIQNMQLAKYQNNWQTAENEKAYARSVEMWNMQNQYNSPTAQMSRLRQAGLNPNL 81
A + N NM++AKYQ WQ ENEKAY RS++MWN+QN+YNSPT QM+R+R AGLNPNL
Sbjct 27 AVQDTNKANMEIAKYQAQWQQQENEKAYQRSLKMWNLQNEYNSPTQQMARIRAAGLNPNL 86
Query 82 VYGSGVTGNSAGSAPQYQPAKIQRATMEPYRGWNLGLSDAASMYMAMRQNKAQVENMEAQ 141
VYG+GVTGNSAGS PQY+PAK TM+ YRGWNLG+SDA S Y+A R KAQV+NMEAQ
Sbjct 87 VYGNGVTGNSAGSTPQYEPAKFNAPTMQAYRGWNLGISDATSQYLAYRTVKAQVDNMEAQ 146
Query 142 NKLIKEQARTEGIRQGNIAMSTARSGFDLNLARELRNVSIDRAIAEKNLSEASAAGAWTG 201
N LI++Q TE RQ NIA ST+RS FDLN+A+EL++VS+ AIAE N +A AA WT
Sbjct 147 NSLIRQQTATEATRQANIAASTSRSEFDLNMAKELKDVSVSSAIAEMNQKQAVAAQGWTK 206
Query 202 ANQKVLQYELDRTLFDNKIKLSNAEYSTAMEALRKLRQDNDINAFRYSMERVFGNSSDVK 261
AN++V+QYELD+ LFDNKIKL+N +Y A++++R+L QDNDINAFRY +E++ G K
Sbjct 207 ANREVVQYELDKALFDNKIKLNNEKYLKALQSVRQLTQDNDINAFRYRIEKITG-----K 261
Query 262 DVASELVKRMGLLLMRPDIKELDQMFNPK 290
+ + R L ++ P+ + D++FNPK
Sbjct 262 GPLLDDLIRRLLTILAPN--DFDRLFNPK 288
>gi|639237431|ref|WP_024568108.1| hypothetical protein [Elizabethkingia anophelis]
Length=287
Score = 76.6 bits (187), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 48/139 (35%), Positives = 74/139 (53%), Gaps = 11/139 (8%)
Query 39 NWQTAENEKAYARSVEMWNMQNQYNSPTAQMSRLRQAGLNPNLVYGSGVTGNSAGSAPQY 98
N + A +A+A ++MWN N+YN+P AQM RL++AGLNPNL+YG G TGNS+
Sbjct 23 NKKIARENRAFA--LDMWNRNNEYNTPLAQMQRLKEAGLNPNLMYGQGTTGNSSS----- 75
Query 99 QPAKIQRATMEPYRGWNLGLSDAASMYMAMRQNKAQVENMEAQNKLIKEQARTEGIRQGN 158
PAK A Y+ L +AA ++ + N+ ++ ++Q +L QA+ N
Sbjct 76 -PAKADGANPTQYK---LNFLEAAQLHQQQKLNEQSIQLQKSQTELNHAQAQKVNAETAN 131
Query 159 IAMSTARSGFDLNLARELR 177
+T + + RE R
Sbjct 132 THANTLNTQETMQFNRESR 150
>gi|494610273|ref|WP_007368519.1| hypothetical protein [Prevotella multiformis]
gi|324988545|gb|EGC20508.1| hypothetical protein HMPREF9141_0987 [Prevotella multiformis
DSM 16608]
Length=437
Score = 70.5 bits (171), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 57/159 (36%), Positives = 80/159 (50%), Gaps = 25/159 (16%)
Query 3 IGAIVGGLGSLAGSMIGAN----AQRQANIQNMQLAKY--QNNWQTAENEKAYARSVEMW 56
IG+ + G SL G + G + AQ +AN N+Q+A+ QN +Q + + A+ MW
Sbjct 69 IGSAISGGVSLLGGLFGGHSNKTAQDRANETNLQIAREANQNQYQMFQEQNAFNE--RMW 126
Query 57 NMQNQYNSPTAQMSRLRQAGLNPNLVYGSGVTGNS-----AGSAPQYQPAKIQRATMEPY 111
N NQYNSP AQM R AG+NP + G+ TGN+ + APQ A++ AT
Sbjct 127 NQMNQYNSPAAQMQRYTDAGINPYIAAGNVQTGNAQSALQSAPAPQQHVAQVMPAT---- 182
Query 112 RGWNLGLSDAASMYMAMRQNKAQVENMEAQNKLIKEQAR 150
G+ DA A N V + AQN+L QA+
Sbjct 183 -----GMGDAVQNSFAQIGN---VISQFAQNQLALAQAK 213
>gi|575094659|emb|CDL66002.1| unnamed protein product [uncultured bacterium]
Length=204
Score = 61.2 bits (147), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 37/90 (41%), Positives = 49/90 (54%), Gaps = 10/90 (11%)
Query 56 WNMQNQYNSPTAQMSRLRQAGLNPNLVYGSG-VTGNSAGSAPQYQPAKIQRATMEPYRGW 114
W+M N YN+P QM RL+ AGLNPNLVYGSG VTGN+ SAP + + G
Sbjct 35 WDMANDYNNPINQMKRLQAAGLNPNLVYGSGSVTGNTT-SAPALTGGQYSYGLDKLVTGG 93
Query 115 NLGLSDAASMYMAMRQNKAQVENMEAQNKL 144
N ++ +N A + N +AQ +L
Sbjct 94 N--------KVASLLKNSADIANTQAQTQL 115
>gi|490418711|ref|WP_004291034.1| hypothetical protein [Bacteroides eggerthii]
gi|217986638|gb|EEC52972.1| hypothetical protein BACEGG_02723 [Bacteroides eggerthii DSM
20697]
Length=368
Score = 62.4 bits (150), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 48/131 (37%), Positives = 62/131 (47%), Gaps = 18/131 (14%)
Query 38 NNWQTAENEKAYARSVEMWNMQNQYNSPTAQMSRLRQAGLNPNLVYGSGVTG------NS 91
N W+ E+ KAY EMWN QN+YN P+AQ +RL AGLNP ++ G G +
Sbjct 77 NAWKLYEDNKAY--QTEMWNKQNEYNDPSAQRARLEAAGLNPYMMMNGGSAGVAGSVSGT 134
Query 92 AGSAPQY-QPAK--IQRATMEP----YRGWNLGLSDAASMYMAMRQ---NKAQVENMEAQ 141
GSAP P+ +Q T P Y G GL A M Q AQ +N+ +
Sbjct 135 QGSAPSAGSPSAQGVQPPTATPYSADYSGVMQGLGHAIDTIMTGSQRNIQNAQADNLRIE 194
Query 142 NKLIKEQARTE 152
K I +A E
Sbjct 195 GKYIASKAIAE 205
>gi|494822881|ref|WP_007558289.1| hypothetical protein [Bacteroides plebeius]
gi|198272097|gb|EDY96366.1| hypothetical protein BACPLE_00802 [Bacteroides plebeius DSM 17135]
Length=344
Score = 60.8 bits (146), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 38/103 (37%), Positives = 62/103 (60%), Gaps = 5/103 (5%)
Query 30 NMQLAKYQNNWQTAENEKAYARSVEMWNMQNQYNSPTAQMSRLRQAGLNPNLVYGSGVTG 89
N+Q+A+ N + + E+ + +MWN +N+YNS ++Q RL +AGLNP ++ G G
Sbjct 47 NIQIAQMSNEYNREQLERQIEQEWDMWNAENEYNSASSQRKRLEEAGLNPYMMMDGGSAG 106
Query 90 N-SAGSAPQYQPA---KIQRATMEPYRGWNL-GLSDAASMYMA 127
+ S+ ++P QPA ++Q ATM+P L GL AS ++A
Sbjct 107 SASSMTSPAAQPAVVPQMQGATMQPADMSGLSGLRGIASEFIA 149
>gi|575094344|emb|CDL65728.1| unnamed protein product [uncultured bacterium]
Length=368
Score = 59.7 bits (143), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 70/266 (26%), Positives = 116/266 (44%), Gaps = 52/266 (20%)
Query 2 PIGAIVGGLGSLAG--SMIGANAQRQANIQNMQLAKYQNNWQTAENEKAYARSVEMWNMQ 59
P+G I GLG + G M G ++ ++ + M+ WQ+ E +K ++MWN
Sbjct 19 PLGFISSGLGIVDGVAGMFGQSSDQRFALGQME-------WQSQEAQKQRDFQLDMWNRN 71
Query 60 NQYNSPTAQMSRLRQAGLNP--NLVYGSGVTGNSAGSAPQ---YQPAKIQRATMEPYRGW 114
N+YN P QM RL +AG+NP ++ S +GNS+ S P PA +++ P
Sbjct 72 NEYNKPDEQMKRLEEAGINPWQSMGNSSVASGNSSLSQPSGFVPSPAHAASSSLSP---- 127
Query 115 NLGLSDAASMYMAMRQ-NKA-------------QVENMEAQNKLIKEQARTEGIRQGNIA 160
LGL AA + + Q NKA ++E + A+ + QA + I N+
Sbjct 128 -LGL--AADVLGKIAQANKAGADTNRVNTLLQTELEKLIAEKNFVSLQAARQAIENSNLP 184
Query 161 MSTARSGFDLNLARELRNVS-------IDRAIAEKNLSEASAAGAWTGANQKVLQYELDR 213
+ L EL + I+ I+ NL+ AA L Y+
Sbjct 185 HIMKKQLDKLTTEIELNKIGKEEVQERINEIISRSNLNNQQAA----------LVYKYGE 234
Query 214 TLFDNKIKLSNAEYSTAMEALRKLRQ 239
+ ++ I +NA+ + + E + LR+
Sbjct 235 RIQESIIAKNNADANRSNEEAKDLRE 260
>gi|647452992|ref|WP_025792810.1| hypothetical protein [Prevotella histicola]
Length=424
Score = 54.3 bits (129), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 45/132 (34%), Positives = 69/132 (52%), Gaps = 17/132 (13%)
Query 22 AQRQANIQNMQLAKYQN--NWQTAENEKAYARSVEMWNMQNQYNSPTAQMSRLRQAGLNP 79
+ R+ N N+Q+A+ N N+Q + A+ +M++ N YN+P+AQM R +AG+NP
Sbjct 25 SNRKTNQTNLQIARETNQMNYQLFQESNAFNE--KMYHEANAYNTPSAQMQRYAEAGINP 82
Query 80 NLVYGSGVTGNSAGSAPQYQPA-KIQRATMEPYRGWNLGLSDAASMYMAMRQNKAQVENM 138
+ G+ TGN+ SA Q PA ++ A M+P G+ A S + N
Sbjct 83 YIAAGNVQTGNTT-SALQSAPAPQMHAAQMQPDVNMANGMMSAGS-----------IINQ 130
Query 139 EAQNKLIKEQAR 150
AQN+L QAR
Sbjct 131 YAQNELALAQAR 142
Lambda K H a alpha
0.313 0.126 0.349 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1498703630544