bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-31_CDS_annotation_glimmer3.pl_2_2
Length=76
Score E
Sequences producing significant alignments: (Bits) Value
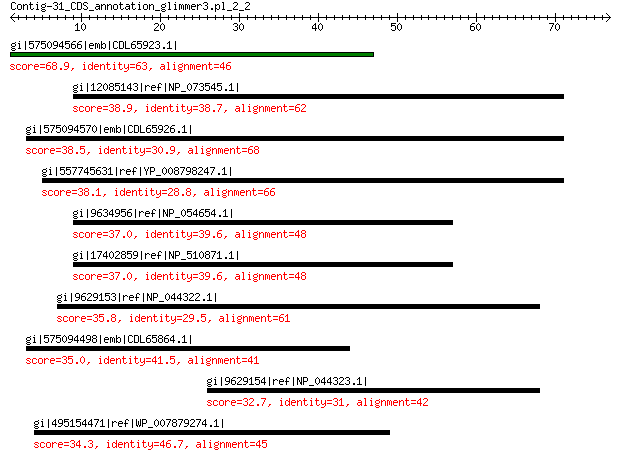
gi|575094566|emb|CDL65923.1| unnamed protein product 68.9 3e-13
gi|12085143|ref|NP_073545.1| nonstructural protein 38.9 0.025
gi|575094570|emb|CDL65926.1| unnamed protein product 38.5 0.037
gi|557745631|ref|YP_008798247.1| packaging protein 38.1 0.060
gi|9634956|ref|NP_054654.1| nonstructural protein 37.0 0.14
gi|17402859|ref|NP_510871.1| nonstructural protein 37.0 0.15
gi|9629153|ref|NP_044322.1| nonstructural protein 35.8 0.39
gi|575094498|emb|CDL65864.1| unnamed protein product 35.0 0.70
gi|9629154|ref|NP_044323.1| nonstructural protein 32.7 3.0
gi|495154471|ref|WP_007879274.1| hypothetical protein 34.3 5.2
>gi|575094566|emb|CDL65923.1| unnamed protein product [uncultured bacterium]
Length=86
Score = 68.9 bits (167), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 29/46 (63%), Positives = 38/46 (83%), Gaps = 0/46 (0%)
Query 1 MTEINLEVAVRNFREGARKNPQIGAFPSDYELHHVGYFDDETGQVL 46
MTE+N E A+RNF+ GA++N QI A P DYELH VG++DDETG+V+
Sbjct 21 MTEVNQESAIRNFKIGAKQNAQISACPEDYELHLVGFWDDETGKVV 66
>gi|12085143|ref|NP_073545.1| nonstructural protein [Bdellovibrio phage phiMH2K]
gi|75089166|sp|Q9G052.1|C_BPPHM RecName: Full=Protein VP5 [Bdellovibrio phage phiMH2K]
gi|12017991|gb|AAG45347.1|AF306496_8 Vp5 [Bdellovibrio phage phiMH2K]
Length=84
Score = 38.9 bits (89), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 24/64 (38%), Positives = 40/64 (63%), Gaps = 9/64 (14%)
Query 9 AVRNFREGARKNPQ--IGAFPSDYELHHVGYFDDETGQVLPVSSNVLETGNHIVIRAEEV 66
A R+F++ A K+PQ + P D++L H+G +DD+TG++ P L+T H V +A ++
Sbjct 28 AERSFQQLA-KDPQSTVANHPEDFDLFHLGEYDDQTGKLTP-----LDTPEHCV-KAIDL 80
Query 67 FKNQ 70
K Q
Sbjct 81 IKQQ 84
>gi|575094570|emb|CDL65926.1| unnamed protein product [uncultured bacterium]
Length=97
Score = 38.5 bits (88), Expect = 0.037, Method: Compositional matrix adjust.
Identities = 21/68 (31%), Positives = 38/68 (56%), Gaps = 1/68 (1%)
Query 3 EINLEVAVRNFREGARKNPQIGAFPSDYELHHVGYFDDETGQVLPVSSNVLETGNHIVIR 62
E N E A RNF +N + A D+EL+ +GYF+DE G ++ S + + + V+R
Sbjct 23 EPNEESAKRNFLFRISQNSFLKAHAKDFELYFLGYFEDELGMLVSASRD-FDRSSIFVLR 81
Query 63 AEEVFKNQ 70
+ + +++
Sbjct 82 GDSLVQSE 89
>gi|557745631|ref|YP_008798247.1| packaging protein [Marine gokushovirus]
gi|530695344|gb|AGT39901.1| packaging protein [Marine gokushovirus]
Length=87
Score = 38.1 bits (87), Expect = 0.060, Method: Compositional matrix adjust.
Identities = 19/66 (29%), Positives = 32/66 (48%), Gaps = 3/66 (5%)
Query 5 NLEVAVRNFREGARKNPQIGAFPSDYELHHVGYFDDETGQVLPVSSNVLETGNHIVIRAE 64
N +A+R F + A + QI P DY L +G F+ TG++ P + + R +
Sbjct 24 NDAIALRQFADMANEETQIAKNPEDYSLWRIGTFETTTGELTPEEPTCIAKAHE---RCD 80
Query 65 EVFKNQ 70
+ KN+
Sbjct 81 TIQKNK 86
>gi|9634956|ref|NP_054654.1| nonstructural protein [Chlamydia phage 2]
gi|47566148|ref|YP_022486.1| nonstructural protein [Chlamydia phage 3]
gi|77020122|ref|YP_338245.1| putative non-structural protein [Chlamydia phage 4]
gi|7406596|emb|CAB85596.1| nonstructural protein [Chlamydia phage 2]
gi|47522483|emb|CAD79484.1| nonstructural protein [Chlamydia phage 3]
gi|59940021|gb|AAX12550.1| putative non-structural protein [Chlamydia phage 4]
Length=84
Score = 37.0 bits (84), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 19/52 (37%), Positives = 28/52 (54%), Gaps = 4/52 (8%)
Query 9 AVRNFREGARKNP---QIGAFPSDYELHHVGYFDDETGQVLPVS-SNVLETG 56
A+R F + ++P Q A P DY L+ +G +DD TG +P+ L TG
Sbjct 26 AIRAFSDMVNEDPTKNQFAAHPEDYILYEIGSYDDSTGTFIPLDVPKALGTG 77
>gi|17402859|ref|NP_510871.1| nonstructural protein [Guinea pig Chlamydia phage]
Length=84
Score = 37.0 bits (84), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 19/52 (37%), Positives = 28/52 (54%), Gaps = 4/52 (8%)
Query 9 AVRNFREGARKNP---QIGAFPSDYELHHVGYFDDETGQVLPVS-SNVLETG 56
A+R F + ++P Q A P DY L+ +G +DD TG +P+ L TG
Sbjct 26 AIRAFSDMVNEDPSKNQFAAHPEDYILYEIGSYDDSTGNFIPLDVPKALGTG 77
>gi|9629153|ref|NP_044322.1| nonstructural protein [Chlamydia phage 1]
gi|139921|sp|P19183.1|C_BPCHP RecName: Full=Protein ORF5 [Chlamydia phage 1]
gi|93812|pir||JU0349 11.5K protein - Chlamydophila psittaci phage Chp1
gi|217772|dbj|BAA00513.1| unnamed protein product [Chlamydia phage 1]
Length=96
Score = 35.8 bits (81), Expect = 0.39, Method: Compositional matrix adjust.
Identities = 18/64 (28%), Positives = 33/64 (52%), Gaps = 8/64 (13%)
Query 7 EVAVRNFREGARKNPQ---IGAFPSDYELHHVGYFDDETGQVLPVSSNVLETGNHIVIRA 63
E A+R FR+ + + +P D++ ++GYFD + G+ PV + ++ +I A
Sbjct 28 EAAIRWFRDVVMDSDSKNILHRYPEDFDFCYIGYFDKDKGRFYPVDAGIV-----TIINA 82
Query 64 EEVF 67
E F
Sbjct 83 GEFF 86
>gi|575094498|emb|CDL65864.1| unnamed protein product [uncultured bacterium]
Length=92
Score = 35.0 bits (79), Expect = 0.70, Method: Compositional matrix adjust.
Identities = 17/42 (40%), Positives = 27/42 (64%), Gaps = 1/42 (2%)
Query 3 EINLEVAVRNFREGAR-KNPQIGAFPSDYELHHVGYFDDETG 43
E N ++A+R F+ + KN I ++P+D+ L VG FD +TG
Sbjct 21 EQNDKLAIRVFQSIYKDKNSTISSYPADFSLFAVGSFDTDTG 62
>gi|9629154|ref|NP_044323.1| nonstructural protein [Chlamydia phage 1]
gi|217773|dbj|BAA00514.1| unnamed protein product [Chlamydia phage 1]
Length=58
Score = 32.7 bits (73), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 13/42 (31%), Positives = 24/42 (57%), Gaps = 5/42 (12%)
Query 26 FPSDYELHHVGYFDDETGQVLPVSSNVLETGNHIVIRAEEVF 67
+P D++ ++GYFD + G+ PV + ++ +I A E F
Sbjct 12 YPEDFDFCYIGYFDKDKGRFYPVDAGIV-----TIINAGEFF 48
>gi|495154471|ref|WP_007879274.1| hypothetical protein [Herbaspirillum sp. CF444]
gi|398098223|gb|EJL88510.1| hypothetical protein PMI16_02154 [Herbaspirillum sp. CF444]
Length=313
Score = 34.3 bits (77), Expect = 5.2, Method: Composition-based stats.
Identities = 21/51 (41%), Positives = 26/51 (51%), Gaps = 6/51 (12%)
Query 4 INLEVAVRNFREGARKNPQIGAF------PSDYELHHVGYFDDETGQVLPV 48
IN E+ VR + GAR I AF Y ++ G F ETGQ+LPV
Sbjct 237 INNEIIVRTWGNGARDQLPIQAFFYTAEAGKRYAMNDQGLFYKETGQLLPV 287
Lambda K H a alpha
0.314 0.132 0.369 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 442533451452