bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-31_CDS_annotation_glimmer3.pl_2_4
Length=259
Score E
Sequences producing significant alignments: (Bits) Value
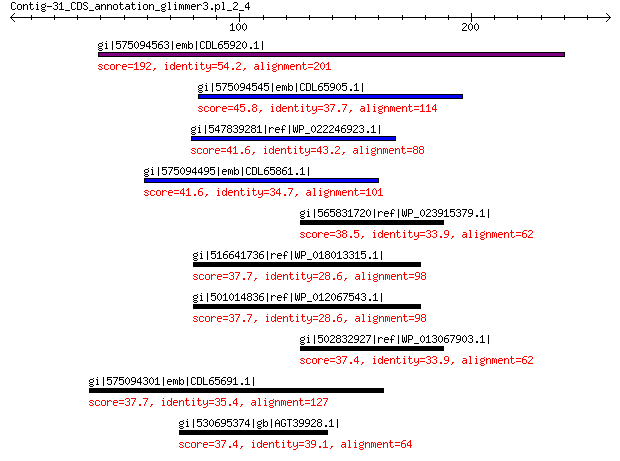
gi|575094563|emb|CDL65920.1| unnamed protein product 192 9e-57
gi|575094545|emb|CDL65905.1| unnamed protein product 45.8 0.016
gi|547839281|ref|WP_022246923.1| putative minor capsid protein 41.6 0.28
gi|575094495|emb|CDL65861.1| unnamed protein product 41.6 0.28
gi|565831720|ref|WP_023915379.1| phospholipase 38.5 2.4
gi|516641736|ref|WP_018013315.1| purine nucleoside phosphorylase 37.7 5.6
gi|501014836|ref|WP_012067543.1| MULTISPECIES: purine nucleoside... 37.7 5.8
gi|502832927|ref|WP_013067903.1| phospholipase 37.4 6.4
gi|575094301|emb|CDL65691.1| unnamed protein product 37.7 7.1
gi|530695374|gb|AGT39928.1| minor capsid protein 37.4 7.2
>gi|575094563|emb|CDL65920.1| unnamed protein product [uncultured bacterium]
Length=242
Score = 192 bits (489), Expect = 9e-57, Method: Compositional matrix adjust.
Identities = 109/211 (52%), Positives = 138/211 (65%), Gaps = 10/211 (5%)
Query 39 GSALGTGMGLVGSAKGLYDSFNNTSLKNQMAYDQFKSELDYQYWSRKMSNRHTLEVGDLR 98
GS +G +G A GLY+ S K Q ++++L + W +MSNRH LEVGDLR
Sbjct 24 GSGIGDILGFGSDALGLYNDLTGNSAKMQKELMAYQAQLQNESWKYQMSNRHQLEVGDLR 83
Query 99 QAGLNPILSANSAGSVASAIPNGAIPETSS-----QQSAAGAAREANRINAMI--GESTS 151
AGLNPILSANSAGSVA+ IPNGA+ ++ S + SAA A + A ++ +++ ST
Sbjct 84 NAGLNPILSANSAGSVAAGIPNGALADSDSARYGARSSAALARQNAAQVASLVQTNASTQ 143
Query 152 AKNLADAQASIMNAETGRMVGIASARRANAEAGLAGTRMSNELAYPNNQPSLFKYINSGK 211
A+N A+A+A +MNA++ RM IA A R NAEAG A R NE YP+NQP FKY NS K
Sbjct 144 ARNEAEAKALLMNAQSNRMSAIAGANRNNAEAGYAAVRSKNESLYPSNQPLPFKYFNSAK 203
Query 212 QLV---EDLFDRNYGLPSNASPARRQRYEVF 239
+V ED DR YGLPSNASP RR+RYEVF
Sbjct 204 GMVDSLEDFLDRRYGLPSNASPERRRRYEVF 234
>gi|575094545|emb|CDL65905.1| unnamed protein product [uncultured bacterium]
Length=325
Score = 45.8 bits (107), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 43/123 (35%), Positives = 59/123 (48%), Gaps = 17/123 (14%)
Query 82 WSRKMSNR-HTLEVGDLRQAGLNPILSANSAGSVASAIPNGAIPE--TSSQQSAAGAARE 138
W MSN H EV DL+ AGLNP+LSA G +A+ +GA + TSS A+
Sbjct 70 WQSYMSNTAHQREVADLKAAGLNPVLSA--MGGNGAAVTSGATAQGYTSSGGQASADTSA 127
Query 139 ANRINAMIGESTSAKNLADAQASIMNAETGRMVGIA------SARRANAEAGLAGTRMSN 192
+ ++G L +AQ SI N T + ++ SA R A+ G AGT S
Sbjct 128 TAALVGLLGS------LLNAQTSIANTATNAVANLSVADKYTSATRYAADVGYAGTSYSA 181
Query 193 ELA 195
+A
Sbjct 182 NVA 184
>gi|547839281|ref|WP_022246923.1| putative minor capsid protein [Clostridium sp. CAG:306]
gi|524476581|emb|CDC18646.1| putative minor capsid protein [Clostridium sp. CAG:306]
Length=236
Score = 41.6 bits (96), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 38/101 (38%), Positives = 52/101 (51%), Gaps = 15/101 (15%)
Query 79 YQYWSRKMSNRHTLEVGDLRQAGLNPILSANSAGSVASAIPNGAI-----PE--TSSQQS 131
+ + R S H EV DLR AGLNPILSA + GS AS P+G + P+ S ++
Sbjct 39 FDFQERMSSTAHQREVKDLRAAGLNPILSAMN-GSGAST-PSGGMGSINTPDYGASIREG 96
Query 132 AAGAAR------EANRINAMIGESTSAKNLADAQASIMNAE 166
A AA+ E N IN + + DAQ+++M AE
Sbjct 97 VAAAAQLGRTKAETNFINQQAMTEQNKRENFDAQSALMRAE 137
>gi|575094495|emb|CDL65861.1| unnamed protein product [uncultured bacterium]
Length=266
Score = 41.6 bits (96), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 35/103 (34%), Positives = 53/103 (51%), Gaps = 19/103 (18%)
Query 59 FNNTSLKNQMAYDQFKSELDYQYWSRKMSNRHTLEVGDLRQAGLNPILSANSAGSVASAI 118
+N S + QMA+ + R S H EV DL AGLNP+LSA +G+ A
Sbjct 66 YNTQSAREQMAFQE-----------RMSSTAHQREVKDLIAAGLNPVLSAGGSGASA--- 111
Query 119 PNGAI--PETSSQQSAAGAAREANRINAMIGESTSAKNLADAQ 159
P+GA+ ++S + A AA + +NA + +AK++ AQ
Sbjct 112 PSGAMATADSSMMSAKANAALQKRIVNAQL---KNAKDINKAQ 151
>gi|565831720|ref|WP_023915379.1| phospholipase [Rhodobacter capsulatus]
gi|564879628|gb|ETD81316.1| phospholipase [Rhodobacter capsulatus YW1]
Length=220
Score = 38.5 bits (88), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 21/63 (33%), Positives = 39/63 (62%), Gaps = 1/63 (2%)
Query 126 TSSQQSAAGAAREANRINAMIGESTSAKNLADAQASIMNAETGRMVGIASA-RRANAEAG 184
++ +++AAG R ++ +NA + + +A+ LA AQ +++ G M+ + A RRA A AG
Sbjct 77 SAPEEAAAGLTRSSDDLNAFLDQVMTAEGLAAAQIALLGFSQGTMMALQVAPRRAQALAG 136
Query 185 LAG 187
+ G
Sbjct 137 VVG 139
>gi|516641736|ref|WP_018013315.1| purine nucleoside phosphorylase [Sinorhizobium medicae]
Length=265
Score = 37.7 bits (86), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 28/103 (27%), Positives = 50/103 (49%), Gaps = 12/103 (12%)
Query 80 QYWSRKMSNRHTLEVGDLRQAGLNPILSANSAGSVASAIPNGAIPETSSQQSAAGAAREA 139
Y+ R +N + + L++ G++ ++ NSAGS+ +P G++ + + AGA
Sbjct 78 HYYERGDANAMRVPIETLKRIGVDNLILTNSAGSLREDMPPGSVMRIADHIAFAGA---- 133
Query 140 NRINAMIGESTSAK-----NLADAQASIMNAETGRMVGIASAR 177
N +IG + A+ N DA ++ ET +GI AR
Sbjct 134 ---NPLIGVESDARFVGMTNAYDAALAVGMEETAERLGIPLAR 173
>gi|501014836|ref|WP_012067543.1| MULTISPECIES: purine nucleoside phosphorylase [Sinorhizobium]
gi|150398530|ref|YP_001328997.1| purine nucleoside phosphorylase [Sinorhizobium medicae WSM419]
gi|150030045|gb|ABR62162.1| purine nucleotide phosphorylase [Sinorhizobium medicae WSM419]
Length=265
Score = 37.7 bits (86), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 28/103 (27%), Positives = 50/103 (49%), Gaps = 12/103 (12%)
Query 80 QYWSRKMSNRHTLEVGDLRQAGLNPILSANSAGSVASAIPNGAIPETSSQQSAAGAAREA 139
Y+ R +N + + L++ G++ ++ NSAGS+ +P G++ + + AGA
Sbjct 78 HYYERGDANAMRVPIETLKRIGVDNLILTNSAGSLREDMPPGSVMRIADHIAFAGA---- 133
Query 140 NRINAMIGESTSAK-----NLADAQASIMNAETGRMVGIASAR 177
N +IG + A+ N DA ++ ET +GI AR
Sbjct 134 ---NPLIGVESDARFVGMTNAYDAALAVGMEETAERLGIPLAR 173
>gi|502832927|ref|WP_013067903.1| phospholipase [Rhodobacter capsulatus]
gi|294677716|ref|YP_003578331.1| phospholipase/carboxylesterase [Rhodobacter capsulatus SB 1003]
gi|294476536|gb|ADE85924.1| phospholipase/carboxylesterase family protein [Rhodobacter capsulatus
SB 1003]
gi|564637532|gb|ETD01030.1| phospholipase [Rhodobacter capsulatus DE442]
gi|564873772|gb|ETD75615.1| phospholipase [Rhodobacter capsulatus R121]
gi|564878082|gb|ETD79828.1| phospholipase [Rhodobacter capsulatus B6]
gi|564972420|gb|ETE53247.1| phospholipase [Rhodobacter capsulatus Y262]
Length=220
Score = 37.4 bits (85), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 21/63 (33%), Positives = 38/63 (60%), Gaps = 1/63 (2%)
Query 126 TSSQQSAAGAAREANRINAMIGESTSAKNLADAQASIMNAETGRMVGIASA-RRANAEAG 184
++ +++AAG R + +NA + + +A+ LA AQ +++ G M+ + A RRA A AG
Sbjct 77 SAPEEAAAGLTRSSEDLNAFLDQVMAAEGLAAAQIALLGFSQGTMMALQVAPRRAQALAG 136
Query 185 LAG 187
+ G
Sbjct 137 VVG 139
>gi|575094301|emb|CDL65691.1| unnamed protein product [uncultured bacterium]
Length=437
Score = 37.7 bits (86), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 45/150 (30%), Positives = 70/150 (47%), Gaps = 32/150 (21%)
Query 35 LGSIGSALGTGMGLVGSAKGLYDSFNNTSL---KNQMAYDQFKSE-------------LD 78
L I A G+VG+A G+YDS SL +Q +++ ++E D
Sbjct 7 LNPINDAGQAISGVVGTATGIYDSV--ASLWGGSSQQKFERHQAEDARNFTHQENALQRD 64
Query 79 Y--QYWSRKMSNRHTLEVGD---LRQAGLNPILSANSAGSVASAIP--NGAIPETSSQQS 131
+ Q W K +N + + L QAG+NP ++ ++AG+ S P G+ T+ Q
Sbjct 65 FARQMW--KDTNDYNTPIAQKQRLEQAGMNPYVNMDNAGTAQSVTPAVGGSAGATTGTQG 122
Query 132 AAGAAREANRINAMIGESTSAKNLADAQAS 161
AA A+ I S +AKNLA+A+ S
Sbjct 123 AAQIAQ-----GNAIASSQAAKNLAEAERS 147
>gi|530695374|gb|AGT39928.1| minor capsid protein [Marine gokushovirus]
Length=249
Score = 37.4 bits (85), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 25/66 (38%), Positives = 35/66 (53%), Gaps = 2/66 (3%)
Query 74 KSELDY-QYWSRKMSNR-HTLEVGDLRQAGLNPILSANSAGSVASAIPNGAIPETSSQQS 131
KS+ D Q ++ +MSN +T + DLR AGLNPIL+A + + G P S
Sbjct 95 KSQFDQSQDFNERMSNTAYTRGIADLRNAGLNPILAATRGVTSTPTVSGGPAPTGSGTGG 154
Query 132 AAGAAR 137
+ AAR
Sbjct 155 SGTAAR 160
Lambda K H a alpha
0.313 0.128 0.365 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1192060985796