bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-33_CDS_annotation_glimmer3.pl_2_1
Length=174
Score E
Sequences producing significant alignments: (Bits) Value
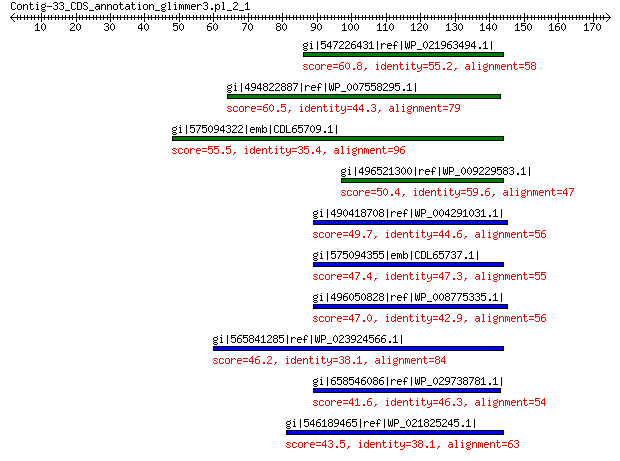
gi|547226431|ref|WP_021963494.1| predicted protein 60.8 5e-08
gi|494822887|ref|WP_007558295.1| hypothetical protein 60.5 8e-08
gi|575094322|emb|CDL65709.1| unnamed protein product 55.5 3e-06
gi|496521300|ref|WP_009229583.1| hypothetical protein 50.4 2e-04
gi|490418708|ref|WP_004291031.1| hypothetical protein 49.7 3e-04
gi|575094355|emb|CDL65737.1| unnamed protein product 47.4 0.001
gi|496050828|ref|WP_008775335.1| hypothetical protein 47.0 0.002
gi|565841285|ref|WP_023924566.1| hypothetical protein 46.2 0.004
gi|658546086|ref|WP_029738781.1| hypothetical protein 41.6 0.019
gi|546189465|ref|WP_021825245.1| hypothetical protein 43.5 0.032
>gi|547226431|ref|WP_021963494.1| predicted protein [Prevotella sp. CAG:1185]
gi|524103383|emb|CCY83995.1| predicted protein [Prevotella sp. CAG:1185]
Length=498
Score = 60.8 bits (146), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 32/58 (55%), Positives = 38/58 (66%), Gaps = 2/58 (3%)
Query 86 QFKGLLKYINIRDYQLFSKRLRKYLSKKIGKYEKIHSYVVSEYSPKTLRPHFHILFFF 143
G L Y + RD QLF KR+RK LSK EKI Y+VSEY PKT R H+H+LFF+
Sbjct 125 NLDGYLSYTSKRDAQLFLKRVRKNLSKYSD--EKIRYYIVSEYGPKTFRAHYHVLFFY 180
>gi|494822887|ref|WP_007558295.1| hypothetical protein [Bacteroides plebeius]
gi|198272100|gb|EDY96369.1| hypothetical protein BACPLE_00805 [Bacteroides plebeius DSM 17135]
Length=545
Score = 60.5 bits (145), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 35/79 (44%), Positives = 45/79 (57%), Gaps = 7/79 (9%)
Query 64 MLPELAESLKKKNNTDANGAFPQFKGLLKYINIRDYQLFSKRLRKYLSKKIGKYEKIHSY 123
M P+L +K+ N N +KG Y++ R+ QLF KRLRKYL K G +KI +
Sbjct 96 MTPQLMNEYQKRVNYRIN-----YKGRFPYLSKRELQLFMKRLRKYLDKYEG--QKIRFF 148
Query 124 VVSEYSPKTLRPHFHILFF 142
EY P + RPHFHIL F
Sbjct 149 ATGEYGPLSFRPHFHILLF 167
>gi|575094322|emb|CDL65709.1| unnamed protein product [uncultured bacterium]
Length=499
Score = 55.5 bits (132), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 34/96 (35%), Positives = 51/96 (53%), Gaps = 7/96 (7%)
Query 48 SPLQRFKDEYFEELVWMLPELAESLKKKNNTDANGAFPQFKGLLKYINIRDYQLFSKRLR 107
S L F +++ +++ + + K + + G GL + RD QLF KRLR
Sbjct 101 SDLHNFDNDFVDKMDYYSDYVINYESKYHKSCVYG-----HGLYALLYYRDIQLFLKRLR 155
Query 108 KYLSKKIGKYEKIHSYVVSEYSPKTLRPHFHILFFF 143
K++ K G EKI Y++ EY K+LRPH+H L FF
Sbjct 156 KHIYKYYG--EKIRFYIIGEYGTKSLRPHWHCLLFF 189
>gi|496521300|ref|WP_009229583.1| hypothetical protein [Prevotella sp. oral taxon 317]
gi|288330571|gb|EFC69155.1| hypothetical protein HMPREF0670_00478 [Prevotella sp. oral taxon
317 str. F0108]
Length=569
Score = 50.4 bits (119), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 28/49 (57%), Positives = 32/49 (65%), Gaps = 2/49 (4%)
Query 97 RDYQLFSKRLRKYLSKKIGKYE--KIHSYVVSEYSPKTLRPHFHILFFF 143
+D Q F KRLR +SK GK E KI YV SEY P TLRPH+H + FF
Sbjct 136 KDIQNFLKRLRFNISKLYGKAESRKIRYYVASEYGPTTLRPHYHGIIFF 184
>gi|490418708|ref|WP_004291031.1| hypothetical protein [Bacteroides eggerthii]
gi|217986635|gb|EEC52969.1| hypothetical protein BACEGG_02720 [Bacteroides eggerthii DSM
20697]
Length=422
Score = 49.7 bits (117), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 25/56 (45%), Positives = 34/56 (61%), Gaps = 1/56 (2%)
Query 89 GLLKYINIRDYQLFSKRLRKYLSKKIGKYEKIHSYVVSEYSPKTLRPHFHILFFFR 144
G L Y+ D QLF KR R Y++K+ K EK+ + + EY P RPH+HIL F +
Sbjct 39 GYLPYLRKFDLQLFFKRFRYYVAKRFPK-EKVRYFAIGEYGPVHFRPHYHILLFLQ 93
>gi|575094355|emb|CDL65737.1| unnamed protein product [uncultured bacterium]
Length=517
Score = 47.4 bits (111), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 26/55 (47%), Positives = 32/55 (58%), Gaps = 2/55 (4%)
Query 89 GLLKYINIRDYQLFSKRLRKYLSKKIGKYEKIHSYVVSEYSPKTLRPHFHILFFF 143
G + Y+ RD QLF KRLRK LSK K+ + + EY P RPH+H L FF
Sbjct 121 GDVPYLRKRDLQLFIKRLRKNLSKYSDA--KVRYFAMGEYGPVHFRPHYHFLLFF 173
>gi|496050828|ref|WP_008775335.1| hypothetical protein [Bacteroides sp. 2_2_4]
gi|229448892|gb|EEO54683.1| hypothetical protein BSCG_01608 [Bacteroides sp. 2_2_4]
Length=497
Score = 47.0 bits (110), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 24/56 (43%), Positives = 34/56 (61%), Gaps = 1/56 (2%)
Query 89 GLLKYINIRDYQLFSKRLRKYLSKKIGKYEKIHSYVVSEYSPKTLRPHFHILFFFR 144
G + Y+ D QLF KRLR Y++K+ EK+ + V EY P RPH+H+L F +
Sbjct 115 GDVPYLRKTDLQLFLKRLRYYVTKQKPS-EKVRYFAVGEYGPVHFRPHYHLLLFLQ 169
>gi|565841285|ref|WP_023924566.1| hypothetical protein [Prevotella nigrescens]
gi|564729906|gb|ETD29850.1| hypothetical protein HMPREF1173_00032 [Prevotella nigrescens
CC14M]
Length=484
Score = 46.2 bits (108), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 32/89 (36%), Positives = 40/89 (45%), Gaps = 5/89 (6%)
Query 60 ELVWMLPELAESLKKKNNTDANGAFPQFKGLLKYINIRDYQLFSKRLRKYLSKKIGKY-- 117
E+VW L + N D + Y D F KRLR LS K+
Sbjct 81 EMVWTSNRLCDEKVIVGNYDFIKVSNSDVQAVAYCCKSDIVKFFKRLRSKLSYYFKKHHI 140
Query 118 ---EKIHSYVVSEYSPKTLRPHFHILFFF 143
EKI +V SEY PKTLRPH+H + +F
Sbjct 141 ITNEKIRYFVCSEYGPKTLRPHYHAIIWF 169
>gi|658546086|ref|WP_029738781.1| hypothetical protein, partial [Elizabethkingia anophelis]
Length=68
Score = 41.6 bits (96), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 25/54 (46%), Positives = 34/54 (63%), Gaps = 7/54 (13%)
Query 89 GLLKYINIRDYQLFSKRLRKYLSKKIGKYEKIHSYVVSEYSPKTLRPHFHILFF 142
GL+ ++ RD+QLF KR RK L KK KI ++V EY +T RPH+H + F
Sbjct 17 GLMS-LDYRDFQLFMKRARK-LQKK-----KISYFLVGEYGSQTHRPHYHAIVF 63
>gi|546189465|ref|WP_021825245.1| hypothetical protein [Prevotella salivae]
gi|544001993|gb|ERK01417.1| hypothetical protein HMPREF9145_2741 [Prevotella salivae F0493]
Length=586
Score = 43.5 bits (101), Expect = 0.032, Method: Compositional matrix adjust.
Identities = 24/67 (36%), Positives = 37/67 (55%), Gaps = 5/67 (7%)
Query 81 NGAFPQFKGLLKYINIRDYQLFSKRLRKYLS----KKIGKYEKIHSYVVSEYSPKTLRPH 136
+G+ P FK L ++ Y L+ + YL+ KK + + ++ SEY+P T RPH
Sbjct 176 DGSIP-FKEWLDDLDTETYDLYYSVYQYYLTDYEKKKESCKQSVRYFICSEYTPTTFRPH 234
Query 137 FHILFFF 143
FH LF+F
Sbjct 235 FHGLFWF 241
Lambda K H a alpha
0.324 0.139 0.425 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 439831946280