bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-33_CDS_annotation_glimmer3.pl_2_2
Length=149
Score E
Sequences producing significant alignments: (Bits) Value
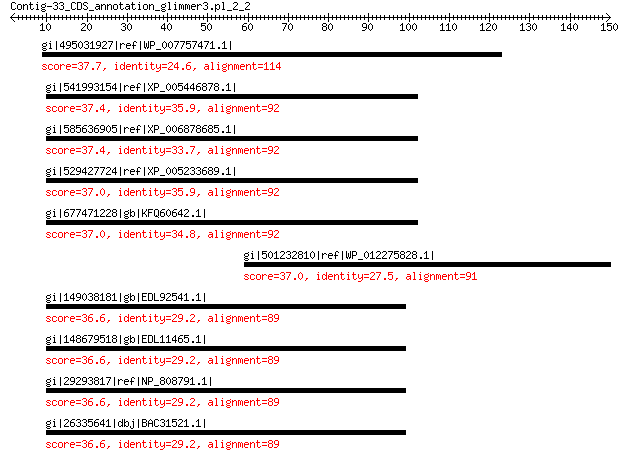
gi|495031927|ref|WP_007757471.1| topoisomerase 37.7 2.0
gi|541993154|ref|XP_005446878.1| PREDICTED: protein VAC14 homolog 37.4 3.0
gi|585636905|ref|XP_006878685.1| PREDICTED: protein VAC14 homolog 37.4 3.1
gi|529427724|ref|XP_005233689.1| PREDICTED: protein VAC14 homolog 37.0 3.4
gi|677471228|gb|KFQ60642.1| Protein VAC14 37.0 3.6
gi|501232810|ref|WP_012275828.1| alkaline phosphatase 37.0 3.7
gi|149038181|gb|EDL92541.1| Vac14 homolog (S. cerevisiae), isofo... 36.6 4.6
gi|148679518|gb|EDL11465.1| Vac14 homolog (S. cerevisiae), isofo... 36.6 4.7
gi|29293817|ref|NP_808791.1| protein VAC14 homolog 36.6 4.8
gi|26335641|dbj|BAC31521.1| unnamed protein product 36.6 5.2
>gi|495031927|ref|WP_007757471.1| topoisomerase [Bacteroides finegoldii]
gi|260620684|gb|EEX43555.1| Toprim domain protein [Bacteroides finegoldii DSM 17565]
Length=675
Score = 37.7 bits (86), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 28/115 (24%), Positives = 53/115 (46%), Gaps = 14/115 (12%)
Query 9 TTLIAML-RTISIVLYLFPLFIKPKNRFARALDSLISLDMRRLEKESKKLRTKGLLYLME 67
T+LI M R +++L+ + +N AR +D L S + R+E +
Sbjct 264 TSLIEMFDRKFRHIVFLYDMDDTGRNESARRMDELSSFHVLRMELPISGAKGD------- 316
Query 68 CLIFRIKNLSDIFPAGRISIDYSPDLPTMMVLFYDGLNKYIQSCSRYYDSLPETS 122
K++SD F +G+ + D+ + +M+ Y ++SC Y++ PE+S
Sbjct 317 ------KDISDYFASGKSAADFQVLITSMLEKLYSQTMMLLKSCEMDYNNPPESS 365
>gi|541993154|ref|XP_005446878.1| PREDICTED: protein VAC14 homolog [Falco cherrug]
Length=986
Score = 37.4 bits (85), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 33/116 (28%), Positives = 52/116 (45%), Gaps = 24/116 (21%)
Query 10 TLIAMLRTISIVLYLFPLFIKPKNRFARALDSLISLDMRRLEKESKKLRTKGLLYLMECL 69
T I M+ I+++ +L+ L+IK + R DSL + +R L ES ++ K L L E
Sbjct 617 TSIGMMTRIAVLKWLYHLYIKTPRKMFRHTDSLFPILLRTLSDESDEVILKDLEVLAEIA 676
Query 70 I--------FRIKNLSDIFP----------AGRIS------IDYSPDLPTMMVLFY 101
++SD+ P AG++S ++ SP PTM FY
Sbjct 677 SSPAGQTEGHGPSDVSDVRPSPVELHVPARAGQLSSSGTKGLECSPSTPTMNSYFY 732
>gi|585636905|ref|XP_006878685.1| PREDICTED: protein VAC14 homolog [Elephantulus edwardii]
Length=785
Score = 37.4 bits (85), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 31/118 (26%), Positives = 49/118 (42%), Gaps = 26/118 (22%)
Query 10 TLIAMLRTISIVLYLFPLFIKPKNRFARALDSLISLDMRRLEKESKKLRTKGLLYLMECL 69
T I M+ I+++ +L+ L+IK + R DSL + ++ L ES ++ K L L E
Sbjct 414 TAIGMMTRIAVLKWLYHLYIKTPRKMFRHTDSLFPILLQTLSDESDEVILKDLEVLAEIA 473
Query 70 I----------------FRIKNLSDIFPA----------GRISIDYSPDLPTMMVLFY 101
R+ ++ PA G ++YSP PTM FY
Sbjct 474 SSPAGQTDDPGPLDGPDLRVSHMELQVPASGRAGPLHAPGTKGLEYSPSTPTMNSYFY 531
>gi|529427724|ref|XP_005233689.1| PREDICTED: protein VAC14 homolog [Falco peregrinus]
Length=871
Score = 37.0 bits (84), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 33/116 (28%), Positives = 52/116 (45%), Gaps = 24/116 (21%)
Query 10 TLIAMLRTISIVLYLFPLFIKPKNRFARALDSLISLDMRRLEKESKKLRTKGLLYLMECL 69
T I M+ I+++ +L+ L+IK + R DSL + +R L ES ++ K L L E
Sbjct 502 TSIGMMTRIAVLKWLYHLYIKTPRKMFRHTDSLFPILLRTLSDESDEVILKDLEVLAEIA 561
Query 70 I--------FRIKNLSDIFP----------AGRIS------IDYSPDLPTMMVLFY 101
++SD+ P AG++S ++ SP PTM FY
Sbjct 562 SSPAGQTEGHGPSDVSDVRPSPVELHVPARAGQLSSSGTKGLECSPSTPTMNSYFY 617
>gi|677471228|gb|KFQ60642.1| Protein VAC14, partial [Pelecanus crispus]
Length=752
Score = 37.0 bits (84), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 32/116 (28%), Positives = 50/116 (43%), Gaps = 24/116 (21%)
Query 10 TLIAMLRTISIVLYLFPLFIKPKNRFARALDSLISLDMRRLEKESKKLRTKGLLYLMECL 69
T I M+ I+++ +L+ L+IK + R DSL + +R L ES ++ K L L E
Sbjct 383 TSIGMMTRIAVLKWLYHLYIKTPRKMFRHTDSLFPILLRTLSDESDEVILKDLEVLAEIA 442
Query 70 I--------FRIKNLSDIFP----------------AGRISIDYSPDLPTMMVLFY 101
+ + SD+ P +G S++ SP PTM FY
Sbjct 443 SSPAGQTEGYSPSDSSDVRPGPVELHVPARAGQLSSSGTKSLECSPSTPTMNSYFY 498
>gi|501232810|ref|WP_012275828.1| alkaline phosphatase [Shewanella halifaxensis]
gi|167622639|ref|YP_001672933.1| alkaline phosphatase [Shewanella halifaxensis HAW-EB4]
gi|167352661|gb|ABZ75274.1| Alkaline phosphatase [Shewanella halifaxensis HAW-EB4]
Length=436
Score = 37.0 bits (84), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 25/92 (27%), Positives = 44/92 (48%), Gaps = 9/92 (10%)
Query 59 TKGLLYLMECLIFRIKNLSDIFPAGRISIDYSPDLP-TMMVLFYDGLNKYIQSCSRYYDS 117
TK L + + L+ L +F A + I +P P M+++ DG+ S RYY
Sbjct 4 TKTLTWTISALV-----LLPLFAAANVDIHDAPSRPKNMIIMIGDGMGPAYTSAYRYYQD 58
Query 118 LPETSPLKSQLRVMYPNLFVGGANTTSVKDAG 149
P+T ++ + ++ L VG A+T + +G
Sbjct 59 NPDTEEIE---QTVFDRLLVGMASTYPARKSG 87
>gi|149038181|gb|EDL92541.1| Vac14 homolog (S. cerevisiae), isoform CRA_a [Rattus norvegicus]
Length=775
Score = 36.6 bits (83), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 26/89 (29%), Positives = 42/89 (47%), Gaps = 9/89 (10%)
Query 10 TLIAMLRTISIVLYLFPLFIKPKNRFARALDSLISLDMRRLEKESKKLRTKGLLYLMECL 69
T I M+ I+++ +L+ L+IK + R DSL + ++ L ES ++ K L L E
Sbjct 404 TTIGMMTRIAVLKWLYHLYIKTPRKMFRHTDSLFPILLQTLSDESDEVVLKDLEVLAEIA 463
Query 70 IFRIKNLSDIFPAGRISIDYSPDLPTMMV 98
PAG+ +PD P + V
Sbjct 464 ---------SSPAGQTDDPGAPDGPDLQV 483
>gi|148679518|gb|EDL11465.1| Vac14 homolog (S. cerevisiae), isoform CRA_d [Mus musculus]
Length=571
Score = 36.6 bits (83), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 26/89 (29%), Positives = 42/89 (47%), Gaps = 9/89 (10%)
Query 10 TLIAMLRTISIVLYLFPLFIKPKNRFARALDSLISLDMRRLEKESKKLRTKGLLYLMECL 69
T I M+ I+++ +L+ L+IK + R DSL + ++ L ES ++ K L L E
Sbjct 424 TTIGMMTRIAVLKWLYHLYIKTPRKMFRHTDSLFPILLQTLSDESDEVVLKDLEVLAEIA 483
Query 70 IFRIKNLSDIFPAGRISIDYSPDLPTMMV 98
PAG+ +PD P + V
Sbjct 484 ---------SSPAGQTDDPGAPDGPDLRV 503
>gi|29293817|ref|NP_808791.1| protein VAC14 homolog [Rattus norvegicus]
gi|81871092|sp|Q80W92.1|VAC14_RAT RecName: Full=Protein VAC14 homolog [Rattus norvegicus]
gi|28864710|gb|AAO48767.1| VAC14 [Rattus norvegicus]
gi|149038182|gb|EDL92542.1| Vac14 homolog (S. cerevisiae), isoform CRA_b [Rattus norvegicus]
Length=783
Score = 36.6 bits (83), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 26/89 (29%), Positives = 42/89 (47%), Gaps = 9/89 (10%)
Query 10 TLIAMLRTISIVLYLFPLFIKPKNRFARALDSLISLDMRRLEKESKKLRTKGLLYLMECL 69
T I M+ I+++ +L+ L+IK + R DSL + ++ L ES ++ K L L E
Sbjct 412 TTIGMMTRIAVLKWLYHLYIKTPRKMFRHTDSLFPILLQTLSDESDEVVLKDLEVLAEIA 471
Query 70 IFRIKNLSDIFPAGRISIDYSPDLPTMMV 98
PAG+ +PD P + V
Sbjct 472 ---------SSPAGQTDDPGAPDGPDLQV 491
>gi|26335641|dbj|BAC31521.1| unnamed protein product [Mus musculus]
Length=558
Score = 36.6 bits (83), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 26/89 (29%), Positives = 42/89 (47%), Gaps = 9/89 (10%)
Query 10 TLIAMLRTISIVLYLFPLFIKPKNRFARALDSLISLDMRRLEKESKKLRTKGLLYLMECL 69
T I M+ I+++ +L+ L+IK + R DSL + ++ L ES ++ K L L E
Sbjct 411 TTIGMMTRIAVLKWLYHLYIKTPRKMFRHTDSLFPILLQTLSDESDEVVLKDLEVLAEIA 470
Query 70 IFRIKNLSDIFPAGRISIDYSPDLPTMMV 98
PAG+ +PD P + V
Sbjct 471 ---------SSPAGQTDDPGAPDGPDLRV 490
Lambda K H a alpha
0.325 0.141 0.405 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 432901228680