bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
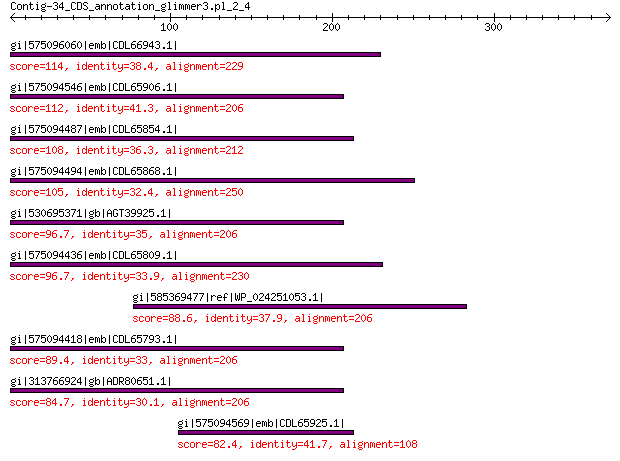
Query= Contig-34_CDS_annotation_glimmer3.pl_2_4
Length=371
Score E
Sequences producing significant alignments: (Bits) Value
gi|575096060|emb|CDL66943.1| unnamed protein product 114 2e-25
gi|575094546|emb|CDL65906.1| unnamed protein product 112 1e-24
gi|575094487|emb|CDL65854.1| unnamed protein product 108 3e-23
gi|575094494|emb|CDL65868.1| unnamed protein product 105 3e-22
gi|530695371|gb|AGT39925.1| replication initiator 96.7 2e-19
gi|575094436|emb|CDL65809.1| unnamed protein product 96.7 2e-19
gi|585369477|ref|WP_024251053.1| hypothetical protein 88.6 4e-17
gi|575094418|emb|CDL65793.1| unnamed protein product 89.4 1e-16
gi|313766924|gb|ADR80651.1| putative replication initiation protein 84.7 2e-15
gi|575094569|emb|CDL65925.1| unnamed protein product 82.4 3e-14
>gi|575096060|emb|CDL66943.1| unnamed protein product [uncultured bacterium]
Length=339
Score = 114 bits (285), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 88/236 (37%), Positives = 117/236 (50%), Gaps = 30/236 (13%)
Query 1 MGCRLDRSRVWADRMLLELKDNDYKALFVTLTYNDRSLPSAWHVGSNYFDGFYedvvplv 60
+GCR+D SR WA+R +LEL+D+D A F T TY++ +P +++ +
Sbjct 56 IGCRIDYSRQWANRCMLELQDHD-SAFFCTFTYDNDHVPISYYADKETGEA--------- 105
Query 61 ldddeeWIaaaagapaTLSIRDTQLFMKrlrktfrdrrlrfflAGEYGPKTHRPHYHAII 120
TL RD QL MKR+RK F D +RFF AGEYG +T RPHYHAII
Sbjct 106 ------------KPSLTLRKRDFQLLMKRIRKHFSDDHIRFFAAGEYGGQTLRPHYHAII 153
Query 121 YGLTLSDFKDCRIKDFNKLGQPRYISKSFERIWGN------GYCVLAPVNWNTCAYVSRY 174
YGL L+D + + Y S S ++ W + G+ V+ V W +CAY +RY
Sbjct 154 YGLHLNDLVPYKTVKEGGVLYTYYNSPSLQKCWLDSDGKPIGFVVVGAVTWESCAYTARY 213
Query 175 TMKKVYKSENSHAYASGQL-PPFCTMSRRPGIGLLHADDLLKKGDKTFIRDIDLNG 229
+KK K E S Y L P F MSR+PGI + D FI L G
Sbjct 214 VLKK-QKGEASTVYQEFNLEPEFTLMSRKPGIARNYYDTHPDLFQSDFINISTLKG 268
>gi|575094546|emb|CDL65906.1| unnamed protein product [uncultured bacterium]
Length=351
Score = 112 bits (280), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 85/218 (39%), Positives = 115/218 (53%), Gaps = 35/218 (16%)
Query 1 MGCRLDRSRVWADRMLLELKDNDYKALFVTLTYNDRSLPSAWHVGSNYFDGFYedvvplv 60
+GCRLD SR WADR++LEL+ + A+FVTLTY++ ++P + DG
Sbjct 62 IGCRLDYSRRWADRLMLELQYHT-AAIFVTLTYSELNVPKHHYQTP---DG--------- 108
Query 61 ldddeeWIaaaagapaTLSIRDTQLFMKrlrktfrdrrlrfflAGEYGPKTHRPHYHAII 120
+L RD QLF KRLRK + D ++RFFL+GEYGPKT RPHYHAII
Sbjct 109 ----------DVNTSYSLDKRDVQLFFKRLRKMYPDTKIRFFLSGEYGPKTFRPHYHAII 158
Query 121 YGLTLS-DFKDCRIKDFNKLGQPRYISKSFERIWGN-----------GYCVLAPVNWNTC 168
+G+ + D R++ + + Y S S ER W G + V+W+TC
Sbjct 159 FGVDFAHDRYVWRVRRADNMFVNYYRSPSLERAWSVYNNDVGDYVPIGNVEFSDVSWHTC 218
Query 169 AYVSRYTMKKVYKSENSHAYASGQLPPFCTMSRRPGIG 206
AYV+RY KK+ + PPF MSR+PGI
Sbjct 219 AYVARYVTKKLTGNLAQFYTTFNLTPPFSLMSRKPGIA 256
>gi|575094487|emb|CDL65854.1| unnamed protein product [uncultured bacterium]
Length=332
Score = 108 bits (269), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 77/213 (36%), Positives = 104/213 (49%), Gaps = 26/213 (12%)
Query 1 MGCRLDRSRVWADRMLLELKDNDYKALFVTLTYNDRSLPSAWHVGSNYFDGFYedvvplv 60
+GCRL +SR WA+R+++E + ++ F+TLTYND LP ++ V
Sbjct 57 VGCRLSKSREWANRVVME-QLYHVESWFLTLTYNDEHLPRSFPVDEA------------- 102
Query 61 ldddeeWIaaaagapaTLSIRDTQLFMKrlrktfrdrrlrfflAGEYGPKTHRPHYHAII 120
TL D Q F+KRLRK + F AGEYG RPHYH +I
Sbjct 103 -------TGEILSVHGTLVKEDLQKFLKRLRKNSGQKLRFFA-AGEYGSLNMRPHYHLLI 154
Query 121 YGLTLSDFKDCRIKDFNKLGQPRYISKSFERIWGNGYCVLAPVNWNTCAYVSRYTMKKVY 180
+GL L D + R + LG Y S E+ W G+ +L V W + AYV+RYTMKK
Sbjct 155 FGLHLEDLQLLRK---SPLGDEYYTSSLLEKCWPFGFHILGRVTWQSAAYVARYTMKKAS 211
Query 181 KSENSHAYASGQL-PPFCTMSRRPGIGLLHADD 212
K + Y L P F MS RPG+ + +D
Sbjct 212 KGYDKDLYKKAALQPEFQVMSNRPGLARQYYED 244
>gi|575094494|emb|CDL65868.1| unnamed protein product [uncultured bacterium]
Length=348
Score = 105 bits (261), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 81/256 (32%), Positives = 124/256 (48%), Gaps = 34/256 (13%)
Query 1 MGCRLDRSRVWADRMLLELKDNDYKALFVTLTYNDRSLPSAWHVGSNYFDGFYedvvplv 60
+GCRL SR WADR +LE + + + F+TLTY+D +LP + + + + Y
Sbjct 68 VGCRLAYSRQWADRCMLESSYHTH-SYFLTLTYDDDNLPLSESINQDTGEINYNA----- 121
Query 61 ldddeeWIaaaagapaTLSIRDTQLFMKr-----lrktfrdrrlrfflAGEYGPKTHRPH 115
TL +D Q F+KR + +++F AGEYG +T RPH
Sbjct 122 ----------------TLVKKDIQDFIKRLRRFCEYNIDDNLHIKYFCAGEYGSQTFRPH 165
Query 116 YHAIIYGLTLSDFKDCRIKDFNKLGQPRYISKSFERIWGNGYCVLAPVNWNTCAYVSRYT 175
YH I+YG ++D K + + G Y S + +++W G+ V+ V W+TCAY +RY
Sbjct 166 YHMILYGFPINDLK---LYKMSLDGYNYYNSATIDKLWKKGFVVIGEVTWDTCAYTARYI 222
Query 176 MKKVYKSENSHAYASGQLPPFCTMSRRPGIGLLHADDLLKKGDKTFIRD-IDLNGKECTR 234
+KK Y S LP F MS +P I + +D DK F D I L KE +
Sbjct 223 LKKQYGSGAQIYKDYNILPEFTCMSTKPAIAREYYED---NKDKIFDSDYIFLGTKEKSI 279
Query 235 EVYLGRAFIRSAAREH 250
++ + F + +E+
Sbjct 280 QMKPPKYFEKLLEKEN 295
>gi|530695371|gb|AGT39925.1| replication initiator [Marine gokushovirus]
Length=316
Score = 96.7 bits (239), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 72/213 (34%), Positives = 111/213 (52%), Gaps = 43/213 (20%)
Query 1 MGCRLDRSRVWADRMLLELKDNDYKALFVTLTYNDRSLPSAWHVGSNYFDGFYedvvplv 60
+GCRL++SR WA R E K + F+TLTYN LP
Sbjct 55 IGCRLEKSRQWALRCTHEAKLYKNNS-FITLTYNSDHLP--------------------- 92
Query 61 ldddeeWIaaaagapaTLSIRDTQLFMKrlrktfrdrrlrfflAGEYGPKTHRPHYHAII 120
+ TL++R QLF+KRLRK + ++ +RF+ GEYG HRPHYHA++
Sbjct 93 ---------LTNNSLPTLNLRHFQLFLKRLRKKYSNKTIRFYHCGEYGDMNHRPHYHALL 143
Query 121 YGLTLSDFKDCRIKDFNKLGQPRYISKSFERIWGN-------GYCVLAPVNWNTCAYVSR 173
+ DF+D ++ +K Q Y S+ + +W + G+ + + +++ AYV+R
Sbjct 144 FN---HDFEDKKLWKIHK-DQNYYTSEVLDGLWTDPKTKSNMGFSTIGDLTFDSAAYVAR 199
Query 174 YTMKKVYKSENSHAYASGQLPPFCTMSRRPGIG 206
Y +KK+ +N+ Y G++P + TMSRRPGIG
Sbjct 200 YCLKKI-TGKNAEDYYQGRVPEYATMSRRPGIG 231
>gi|575094436|emb|CDL65809.1| unnamed protein product [uncultured bacterium]
Length=340
Score = 96.7 bits (239), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 78/236 (33%), Positives = 109/236 (46%), Gaps = 24/236 (10%)
Query 1 MGCRLDRSRVWADRMLLELKDNDYKALFVTLTYNDRSLPSAWHVGSNYFDGFYedvvplv 60
+GCR+ + WA R+ LE + +A FVTLTY D ++P +G
Sbjct 55 IGCRIRAKQDWATRLELEARAYKGRAWFVTLTYRDDTIPLLIRNTGELIEGGVSMWSRGA 114
Query 61 ldddeeWIaaaagapaTLSIRDTQLFMKrlrktfrdrrlrffl-----AGEYGPKTHRPH 115
++ TL++ D F KRLRK AGEYG +T RPH
Sbjct 115 DVPEQI---------NTLNMDDVTKFWKRLRKYQTTEPDMGKELRYFYAGEYGEQTGRPH 165
Query 116 YHAIIYGLTLSDFKDCRIKDFNKLGQPRYISKSFERIWGNGYCVLAPVNWNTCAYVSRYT 175
YHAII+GL + D K ++ N+ Y S E+IWG G +A T YV+ Y
Sbjct 166 YHAIIFGLEIPDLK--KVPGRNQY----YKSAILEKIWGKGNVTIAYSEPGTYNYVAGYV 219
Query 176 MKKVYKSENSHAYASGQLPPFCTMSRRPGIGLLHADDLLKKGDKTFIRD-IDLNGK 230
KK+Y ++ G P+ MSR+PGIG+ + L DK + +D I L GK
Sbjct 220 TKKMYGNDTKEYQNLGLTAPYACMSRKPGIGMPWLEQNL---DKLWEQDYIQLAGK 272
>gi|585369477|ref|WP_024251053.1| hypothetical protein [Escherichia coli]
Length=243
Score = 88.6 bits (218), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 78/240 (33%), Positives = 101/240 (42%), Gaps = 42/240 (18%)
Query 77 TLSIRDTQLFMKrlrktfrdrrlrfflAGEYGPKTHRPHYHAIIYGLTLSDFKDCRIKDF 136
+L RD QLF KRLRK F D +R+F GEYG T RPHYHAI++GL L D +
Sbjct 5 SLCKRDLQLFWKRLRKAFPDDHIRYFACGEYGSTTFRPHYHAIVFGLHLHDLIPVQDIRR 64
Query 137 NKLGQPRYISKSFERIWGN----------------GYCVLAPVNWNTCAYVSRYTMKKVY 180
+G + S+S +R W GY ++ VNW TCAYV+RY +KK
Sbjct 65 GDVGYQYFYSESLQRAWSVVEQKGEYDTPCIRKPIGYVLVGQVNWETCAYVARYVLKKAC 124
Query 181 KSENSHAYASGQLPPFCTMSRRPGIGLLHADDLLK------------------KGDKTFI 222
E P + MSRRPGIG DD + + K F
Sbjct 125 GPEADVYQTFNIQPEYVDMSRRPGIGRQWYDDHPECMEYDTISISTPDGGRKIRPPKYFD 184
Query 223 RDIDLNGKECTREVYLGRAFIRSAAREHMKPVFAAADLVESVQTCINELEAESCTEHEDV 282
+ DL E E+ A R+H A L +S T LE + H +
Sbjct 185 KLFDLEQPELMAEI--------KAKRKHFAEEGKKAKLAQSTMTYEEILETQERVLHNRI 236
>gi|575094418|emb|CDL65793.1| unnamed protein product [uncultured bacterium]
Length=367
Score = 89.4 bits (220), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 68/211 (32%), Positives = 103/211 (49%), Gaps = 20/211 (9%)
Query 1 MGCRLDRSRVWADRMLLELKDNDYK-ALFVTLTYNDRSLPSAWHVGSNYFDGFYedvvpl 59
+ CR+ + WA R EL+ N +K ++F+TLTY++ +P + G
Sbjct 50 LACRIQYAANWAAR--CELETNYHKQSIFLTLTYDEEHVPVLNKETGEIYRGV------- 100
Query 60 vldddeeWIaaaagapaTLSIRDTQLFMKrlrktfrdrrlrffl----AGEYGPKTHRPH 115
+ E++A T+ D Q F+KRLRK L + +GEYG KT RPH
Sbjct 101 --RNPAEYVAGVTLERMTVYKPDVQKFIKRLRKAAEKEGLTDHIMYYLSGEYGDKTGRPH 158
Query 116 YHAIIYGLTLSDFKDCRIKDFNKLGQPRYISKSFERIWGNGYCVLAPVNWNTCAYVSRYT 175
YH I+YGL + D + ++ G R+ S+ + IWG G + V + +C YV+RY
Sbjct 159 YHLIVYGLEVPDAEHI----GSRRGYDRFTSEWLKGIWGMGLIEIGSVTYESCQYVARYV 214
Query 176 MKKVYKSENSHAYASGQLPPFCTMSRRPGIG 206
+KK E +G +P F MS +P IG
Sbjct 215 IKKRKGKEAKEYKDAGIMPEFVQMSLKPAIG 245
>gi|313766924|gb|ADR80651.1| putative replication initiation protein [Uncultured Microviridae]
Length=285
Score = 84.7 bits (208), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 62/217 (29%), Positives = 93/217 (43%), Gaps = 36/217 (17%)
Query 1 MGCRLDRSRVWADRMLLE--LKDNDYKALFVTLTYNDRSLPSAWHVGSNYFDGFYedvvp 58
+GCRLD + +WA R+ E L D+ F+TLTY++ LP W + ++F F + +
Sbjct 9 IGCRLDHAGMWASRIEHESSLYDDSNGNCFITLTYDEEHLPQDWSLDKSHFQKFMKRLRK 68
Query 59 lvldddeeWIaaaagapaTLSIRDTQLFMKrlrktfrdrrlrfflAGEYGPKTHRPHYHA 118
+ G I T G RPHYHA
Sbjct 69 RYPQKIRYYHCGEYGENCRHGIHTTLCP---------------------GCNVGRPHYHA 107
Query 119 IIYGLTLSDFKDCRIKDFNKLGQPRYISKSFERIWGNGYCVLAPVNWNTCAYVSRYTMKK 178
I++ + DF D R+ G P + S + IWG+G+ + + + YV+RY +KK
Sbjct 108 ILFNI---DFHD-RVLVGQSKGIPHFTSDTLTEIWGHGFTQVGDLTAQSAGYVARYALKK 163
Query 179 VYKSENSHAYASGQL---------PPFCTMSRRPGIG 206
V ++ Y S L P + TMSR+PGIG
Sbjct 164 VTGTQAEDHYRSIDLTTGEVTYVRPEYATMSRKPGIG 200
>gi|575094569|emb|CDL65925.1| unnamed protein product [uncultured bacterium]
Length=354
Score = 82.4 bits (202), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 45/108 (42%), Positives = 60/108 (56%), Gaps = 3/108 (3%)
Query 105 GEYGPKTHRPHYHAIIYGLTLSDFKDCRIKDFNKLGQPRYISKSFERIWGNGYCVLAPVN 164
GEYG T RPHYHAI++G +D + K+F Y+SKS IW NG ++ V
Sbjct 160 GEYGDTTFRPHYHAILFGWRPTDLIQFK-KNFQ--NDTLYLSKSLASIWQNGNVMVGDVT 216
Query 165 WNTCAYVSRYTMKKVYKSENSHAYASGQLPPFCTMSRRPGIGLLHADD 212
+C YV+RY +KK ++ G LP F TMSR+PGI + DD
Sbjct 217 PESCRYVARYCLKKATGFDSEIYERLGVLPEFVTMSRKPGIARKYFDD 264
Lambda K H a alpha
0.324 0.139 0.438 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 2277859962564