bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-3_CDS_annotation_glimmer3.pl_2_3
Length=515
Score E
Sequences producing significant alignments: (Bits) Value
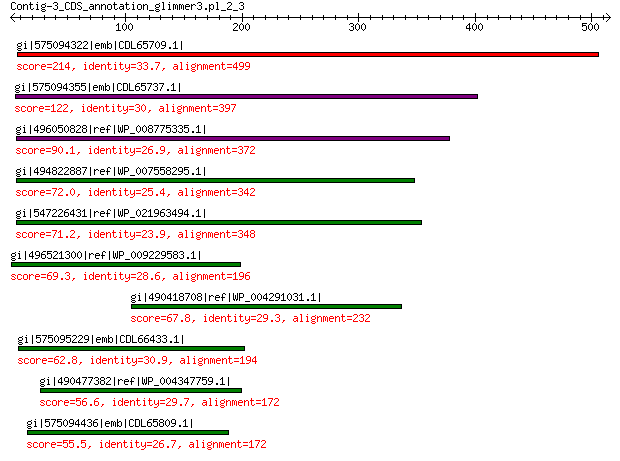
gi|575094322|emb|CDL65709.1| unnamed protein product 214 4e-59
gi|575094355|emb|CDL65737.1| unnamed protein product 122 1e-26
gi|496050828|ref|WP_008775335.1| hypothetical protein 90.1 6e-16
gi|494822887|ref|WP_007558295.1| hypothetical protein 72.0 4e-10
gi|547226431|ref|WP_021963494.1| predicted protein 71.2 6e-10
gi|496521300|ref|WP_009229583.1| hypothetical protein 69.3 3e-09
gi|490418708|ref|WP_004291031.1| hypothetical protein 67.8 6e-09
gi|575095229|emb|CDL66433.1| unnamed protein product 62.8 3e-07
gi|490477382|ref|WP_004347759.1| hypothetical protein 56.6 3e-05
gi|575094436|emb|CDL65809.1| unnamed protein product 55.5 5e-05
>gi|575094322|emb|CDL65709.1| unnamed protein product [uncultured bacterium]
Length=499
Score = 214 bits (545), Expect = 4e-59, Method: Compositional matrix adjust.
Identities = 168/513 (33%), Positives = 250/513 (49%), Gaps = 43/513 (8%)
Query 7 FIKCTSPIAIH--RGSSIFLYPCRKCECCQVSRQKSLSTMLALEESHAKYCYFINPTYND 64
F+KC SP+ + RG + PC KC C +++ SLS L LEE +KYCYF+ TY+D
Sbjct 5 FVKCFSPLVLRDPRGYP-YQVPCGKCIACHNNKRSSLSLKLRLEEYTSKYCYFLTLTYDD 63
Query 65 FNIPAVRVPDDADFGSIAEFEIITP---RLKRDKYFEPYIVDYDVDLEKSIGQLCEQRDE 121
N+P V D EF I P RL+ D + I D+ DL ++ D
Sbjct 64 DNLPLFSVGLDT---CATEFVRIYPYSERLRNDSF----ISDFCSDLHNFDNDFVDKMDY 116
Query 122 YSRLY----SRSHK--FVPHDVIYLLHYPDIQRFIKRFRIYAKRKFNASCRYYVVGEYGT 175
YS S+ HK H + LL+Y DIQ F+KR R + + + R+Y++GEYGT
Sbjct 117 YSDYVINYESKYHKSCVYGHGLYALLYYRDIQLFLKRLRKHIYKYYGEKIRFYIIGEYGT 176
Query 176 NSLRPHWHLLLFFDSDELAQDLERCYHPTDKELRKDPDLRFYNDHDCAECLRTLWKFGFA 235
SLRPHWH LLFF+S L+Q E C + C LR W+FG
Sbjct 177 KSLRPHWHCLLFFNSSSLSQAFEDCVNVGTTS----------RPCSCPRFLRPFWQFGIC 226
Query 236 TSERTNKsayyyvsgyvtstsRFPLCLSALSRPHSLHSRFFGQTLAEEEIQRAIVSQDFE 295
S+RTN AY YVS YV ++ FP L LS + HS GQ L+E+ I AI DF
Sbjct 227 DSKRTNGEAYNYVSSYVNQSANFPKLLVLLSNQKAYHSIQLGQILSEQSIVSAIQKGDFS 286
Query 296 YFRVHYRYSSKGYQIPYALWRSYYSRFFPVFTGLVNMSNEKIFRLFEYWEEIGKLSGYYR 355
+F + + G Y++WRSYYSRFFP FT ++ E+ +R+ +E + L
Sbjct 287 FFERQFYLDTFGAANSYSVWRSYYSRFFPKFTCSSQLTYEQTYRVLTCYETLRDLFDTDS 346
Query 356 VSNQVQWLKNWYYYYYSRPLETIPSPIADVLLNLQSTFDLSEHCLHPEDFSALHDVLYAS 415
V + L +Y+Y++ P D+ L+ ++ + FSAL + AS
Sbjct 347 VGVICRRL--FYHYHFGYP------DYHDIFDFLRFAYNAVLNSKDISLFSALRSCVSAS 398
Query 416 KRFCWLCSWLNMSHKQYFEIWSNFYKYINLSLYKEHYLALENDSKYFRGWFD---RRFIT 472
+ F + ++ YF + +FY+Y++LS + H+ S+Y +++ R I
Sbjct 399 RTFLRAAAMCGLTPTAYFRKYKDFYRYLDLSHLRSHFENCIASSEYSNNYYNIYQLRTIN 458
Query 473 PTAVLEFPLADSDFTNFVENSVSTHDSLIKHRE 505
+ + +DS+F+ F + + IKHR+
Sbjct 459 GNYIYD---SDSEFSRFRVSEQIRFERSIKHRD 488
>gi|575094355|emb|CDL65737.1| unnamed protein product [uncultured bacterium]
Length=517
Score = 122 bits (305), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 119/420 (28%), Positives = 182/420 (43%), Gaps = 56/420 (13%)
Query 5 LPFIKCTSPIAIHRG--SSIFLYPCRKCECCQVSRQKSLSTMLALEESHAKYCYFINPTY 62
PFI+C P + + L PC KC CQ S+ + LE S K+C F TY
Sbjct 6 FPFIRCLEPKRVFNPYLNDWLLVPCGKCRACQCSKASRYKLQIQLEASQHKFCIFGTLTY 65
Query 63 NDFNIPAVR-VP-DDADFGSIAEFEIITPRLKRDKYFEPYIVDYDVDLEKSIGQLCEQRD 120
+ IP + VP +D FG + +E+ D + + +G L
Sbjct 66 ANTYIPRLSLVPYNDKTFGVVNGYEMC-----------------DKETGEYLGYLDSPSY 108
Query 121 EYSRLYSRSHKFVPHDVIYLLHYPDIQRFIKRFRIYAKRKFNASCRYYVVGEYGTNSLRP 180
+ L + H F DV YL D+Q FIKR R + +A RY+ +GEYG RP
Sbjct 109 DVESLLDKLHLF--GDVPYL-RKRDLQLFIKRLRKNLSKYSDAKVRYFAMGEYGPVHFRP 165
Query 181 HWHLLLFFDSDELAQDLERCYHPTDKELRKDPDLRFYNDHD-CAE---------CLRTLW 230
H+H LLFFD + P+ L + PD +Y+ + C+ C+R+ W
Sbjct 166 HYHFLLFFDEIKFTA-------PSGHTLGEFPDWAWYDSQNKCSRSDILSVVEYCIRSSW 218
Query 231 KFGFATSERTNKsayyyvsgyvtstsRFPLCLSALS-RPHSLHSRFFGQTLAEEEIQRAI 289
KFG ++ + A YVS YV+ + P S RP SLHSRF GQ E ++
Sbjct 219 KFGRVDAQYSKGDAAQYVSSYVSGSGSLPKVYQVSSARPFSLHSRFLGQGFLAHECEKVY 278
Query 290 VS--QDFEYFRVHYRYSSKGYQIPYALWRSYYSRFFPVFTGLVNMSNEKIFRLFEYWEEI 347
+ +DF V S+K + LWRS YS F+P G S+ + ++ ++
Sbjct 279 ETPVRDFVKRSVELNGSNKDFN----LWRSCYSVFYPKCKGFTRKSSSERLYTYKLYDTA 334
Query 348 GKLSGYYRVSNQVQWLK------NWYYYYYSRPLETIPSPIADVLLNLQSTFDLSEHCLH 401
+L Y VS+ ++ + +Y Y + + I LL + + +++E L
Sbjct 335 KRLFPY--VSSVIELARETMIHLTFYVYGKQHTVAELDYDIKRYLLYFRDSLNINEVVLQ 392
>gi|496050828|ref|WP_008775335.1| hypothetical protein [Bacteroides sp. 2_2_4]
gi|229448892|gb|EEO54683.1| hypothetical protein BSCG_01608 [Bacteroides sp. 2_2_4]
Length=497
Score = 90.1 bits (222), Expect = 6e-16, Method: Compositional matrix adjust.
Identities = 100/379 (26%), Positives = 151/379 (40%), Gaps = 59/379 (16%)
Query 6 PFIKCTSPIAIHR--GSSIFLYPCRKCECCQVSRQKSLSTMLALEESHAKYCYFINPTYN 63
PF KC P I + PC C+ C +++ + LE AK+ FI TY
Sbjct 5 PFCKCLHPKRIMNPYTKESMVVPCGHCQACTLAKNSRYAFQCDLESYTAKHTLFITLTYA 64
Query 64 DFNIPAVRVPDDADFGSIAEFEIITPRLKRDKYFEPYIVDY-DVDLEKSIGQLCEQRDEY 122
+ IP D + PY D D + + +G DE
Sbjct 65 NRFIPRAMFVDSIE--------------------RPYGCDLIDKETGEILGPADLTEDER 104
Query 123 SRLYSRSHKFVPHDVIYLLHYPDIQRFIKRFRIYA-KRKFNASCRYYVVGEYGTNSLRPH 181
+ L ++ + F DV YL D+Q F+KR R Y K+K + RY+ VGEYG RPH
Sbjct 105 TNLLNKFYLF--GDVPYL-RKTDLQLFLKRLRYYVTKQKPSEKVRYFAVGEYGPVHFRPH 161
Query 182 WHLLLFFDSDELAQDLERCYHPTDKELRKDPDLRFYNDHDCAECLRTLWKFGFATSERTN 241
+HLLLF SDE Q C+E + W FG + +
Sbjct 162 YHLLLFLQSDEALQI-------------------------CSENISKAWTFGRVDCQVSK 196
Query 242 KsayyyvsgyvtstsRFPLCLSALSR-PHSLHSRFFGQTLAEEEIQR--AIVSQDFEYFR 298
YV+ YV S+ P A S P S+HS+ GQ + + ++ ++ ++F
Sbjct 197 GQCSNYVASYVNSSCTIPKVFKASSVCPFSVHSQKLGQGFLDCQREKIYSLTPENF---- 252
Query 299 VHYRYSSKGYQIPYALWRSYYSRFFPVFTGLVNMSNEKIFRLFEYWEEIGKLSGYYRVSN 358
+ G + +WRS YS F+P G V S+ + + ++ L + +
Sbjct 253 IRSSIVLNGKYKEFDVWRSCYSFFYPRCKGFVTKSSRERAYSYSIYDTARLLFPDAKTTF 312
Query 359 QVQWLKNWYYYYYSRPLET 377
+ Y YY+ P ET
Sbjct 313 SLAKEIAIYIYYFHNPKET 331
>gi|494822887|ref|WP_007558295.1| hypothetical protein [Bacteroides plebeius]
gi|198272100|gb|EDY96369.1| hypothetical protein BACPLE_00805 [Bacteroides plebeius DSM 17135]
Length=545
Score = 72.0 bits (175), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 87/370 (24%), Positives = 148/370 (40%), Gaps = 70/370 (19%)
Query 6 PFIKCTSPIAI---HRGSSIFLYPCRKCECCQVSRQKSLSTMLALEESHAKYCYFINPTY 62
PF+ C P I + G + + C+ C C+ R S + E AK F+ T+
Sbjct 4 PFVSCLEPQRIKNKYTGEEMVV-ACKHCVACEQLRNFKYSNLCDFESLTAKKTVFLTLTF 62
Query 63 NDFNIPAVR---------VPDDADFGSIAEFEIITPRLKRDKYFEPYIVDYDVDLEKSIG 113
+D +P R + DAD G ++TP+L + +K +
Sbjct 63 DDKFVPQFRFYKVGDDEYIMRDADTGEYLGRTLMTPQLMNE-------------YQKRVN 109
Query 114 QLCEQRDEYSRLYSRSHKFVPHDVIYLLHYPDIQRFIKRFRIYAKRKFNASCRYYVVGEY 173
+ + L R ++Q F+KR R Y + R++ GEY
Sbjct 110 YRINYKGRFPYLSKR----------------ELQLFMKRLRKYLDKYEGQKIRFFATGEY 153
Query 174 GTNSLRPHWHLLLFFDSDELAQDLERCYHPTDKELRKDPDLRFYNDHDCAEC-------- 225
G S RPH+H+LLF D L + P+ L + P +++ + A C
Sbjct 154 GPLSFRPHFHILLFVDDPSL-------FLPSVHTLGEYP-YPYWSKYQKAHCGKGTLLSK 205
Query 226 ----LRTLWKFGFATSERTNKsa-yyyvsgyvtstsRFPLCLSALS-RPHSLHSRFFGQT 279
+R W FG ++ + + YV+GYV S+ P CL + + S HSRF G+
Sbjct 206 LEYYIRESWPFGGIDAQSVEQGSCSSYVAGYVNSSVPLPSCLKVDAVKSFSQHSRFLGRK 265
Query 280 LAEEEIQRAIVSQDFEYFRVHY--RYSSKGYQIPYALWRSYYSRFFPVFTGLVNMSNEKI 337
+ E+ + + E+ + + R ++ P + S Y P G +S+E+
Sbjct 266 IFGTELIPLLKLKFTEFVQRSFFCRGRYDNFRTPSEMLHSVY----PQCKGFALLSHEQR 321
Query 338 FRLFEYWEEI 347
FR++ W +
Sbjct 322 FRVYTIWSRL 331
>gi|547226431|ref|WP_021963494.1| predicted protein [Prevotella sp. CAG:1185]
gi|524103383|emb|CCY83995.1| predicted protein [Prevotella sp. CAG:1185]
Length=498
Score = 71.2 bits (173), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 83/354 (23%), Positives = 134/354 (38%), Gaps = 47/354 (13%)
Query 6 PFIKCTSPIAIHR--GSSIFLYPCRKCECCQVSRQKSLSTMLALEESHAKYCYFINPTYN 63
P +KC P + + C C+ C R +S + A+EE KYC F TY+
Sbjct 7 PLVKCYHPRHVQNKYTGEVIQVGCGVCKACLKRRADKMSFLCAIEEQSHKYCMFATLTYS 66
Query 64 DFNIPAVRVPDDADFGSIAEFEIITPRLKRDKYFEPYIVDYDVDLEKSIGQLCEQRDEYS 123
+ +P + D + + + ++ K VDYD C D Y
Sbjct 67 NDYVPRMYPEVDNELRLVRWYSYCDRLNEKGKLM---TVDYDY------WHKCPSLDTYV 117
Query 124 RLYSRSHKFVPHDVIYLLHYPDIQRFIKRFRIYAKRKFNASCRYYVVGEYGTNSLRPHWH 183
+ + + + D Q F+KR R + + RYY+V EYG + R H+H
Sbjct 118 LMLTAKCNLDGY--LSYTSKRDAQLFLKRVRKNLSKYSDEKIRYYIVSEYGPKTFRAHYH 175
Query 184 LLLFFDSDELAQDLERCYHPTDKELRKDPDLRFYNDHDCAECLRTLWKFGFATSERTNKs 243
+L F+D + T K + K +R W+FG +
Sbjct 176 VLFFYDEVK-----------TQKVMSK--------------VIRQAWQFGRVDCSLSRGK 210
Query 244 ayyyvsgyvtstsRFPLCLSALS-RPHSLHSRFFGQTL---AEEEIQRAIVSQDFEYFRV 299
YV+ YV P L +S +P S HS F + +EEI + V DF +
Sbjct 211 CNSYVARYVNCNYCLPRFLGDMSTKPFSCHSIRFALGIHQSQKEEIYKGSV-DDF----I 265
Query 300 HYRYSSKGYQIPYALWRSYYSRFFPVFTGLVNMSNEKIFRLFEYWEEIGKLSGY 353
+ G + + WR+ FFP G S+ ++++ + E+ GY
Sbjct 266 YQSGEINGNYVEFMPWRNLSCTFFPKCKGYSRKSDSELWQSYNILREVRSAIGY 319
>gi|496521300|ref|WP_009229583.1| hypothetical protein [Prevotella sp. oral taxon 317]
gi|288330571|gb|EFC69155.1| hypothetical protein HMPREF0670_00478 [Prevotella sp. oral taxon
317 str. F0108]
Length=569
Score = 69.3 bits (168), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 56/209 (27%), Positives = 88/209 (42%), Gaps = 31/209 (15%)
Query 2 ANLLPFIKCTSPIAIHR--GSSIFLYPCRKCECCQVSRQKSLSTMLALEESHAKYCYFIN 59
++L F C P+ +H PC +CE C + S + E KY
Sbjct 3 SDLKIFGNCLCPVHVHNRWTRDEMFVPCGRCEACVNAAASKQSKRVRNEIMQHKYSVMFT 62
Query 60 PTYNDFNIPA-VRVPDDAD------FGSIAEFEIITPRLKRDKYFEPYIVDYDVDLEKSI 112
TYN+ IP R D+ D G AE P YF+ + +DL+ +
Sbjct 63 LTYNNEFIPRWERFLDNNDCPQLRPIGRCAELFPSCPL----NYFDKVTGKWSIDLDTFL 118
Query 113 GQLCEQRDEYSRLYSRSHKFVPHDVIYLLHYPDIQRFIKRFRIYAKRKFNAS----CRYY 168
++ + DE++ +++ K DIQ F+KR R + + + RYY
Sbjct 119 PKI--ENDEHTEVFASCCK------------KDIQNFLKRLRFNISKLYGKAESRKIRYY 164
Query 169 VVGEYGTNSLRPHWHLLLFFDSDELAQDL 197
V EYG +LRPH+H ++FFD L ++
Sbjct 165 VASEYGPTTLRPHYHGIIFFDDASLLSEI 193
>gi|490418708|ref|WP_004291031.1| hypothetical protein [Bacteroides eggerthii]
gi|217986635|gb|EEC52969.1| hypothetical protein BACEGG_02720 [Bacteroides eggerthii DSM
20697]
Length=422
Score = 67.8 bits (164), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 68/236 (29%), Positives = 102/236 (43%), Gaps = 36/236 (15%)
Query 105 DVDLEKSIGQLCEQRDEYSRLYSRSHKF--VPHDVIYLLHYPDIQRFIKRFRIYAKRKF- 161
DV+ + +G+ E RL + H F +P+ L D+Q F KRFR Y ++F
Sbjct 11 DVETGEYLGEADLSIKEIERLQEKFHLFGYLPY-----LRKFDLQLFFKRFRYYVAKRFP 65
Query 162 NASCRYYVVGEYGTNSLRPHWHLLLFFDSDELAQDLERCYHPTDKELRKDPDLRFYNDHD 221
RY+ +GEYG RPH+H+LLF SDE Q
Sbjct 66 KEKVRYFAIGEYGPVHFRPHYHILLFLQSDEALQV------------------------- 100
Query 222 CAECLRTLWKFGFATSERTNKsayyyvsgyvtstsRFPLCLSALSR-PHSLHSRFFGQTL 280
C++ + W FG + + YV+GYV S+ P L+ + P +HS+ GQ
Sbjct 101 CSKVVSEAWPFGRVDCQLSKGKCSSYVAGYVNSSVLVPKVLTLPTLCPFCVHSQKLGQGF 160
Query 281 AEEEIQRAIVSQDFEYFRVHYRYSSKGYQIPYALWRSYYSRFFPVFTGLVNMSNEK 336
+ E + + S E F V G + +WRS Y+ FFP G + S+ +
Sbjct 161 LQSE-RAKVYSLTPEQF-VKRSIVINGRYKEFDVWRSAYAYFFPKCKGFADKSSRE 214
>gi|575095229|emb|CDL66433.1| unnamed protein product [uncultured bacterium]
Length=510
Score = 62.8 bits (151), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 60/209 (29%), Positives = 93/209 (44%), Gaps = 20/209 (10%)
Query 8 IKCTSPIAIHRGSSIFLYPCRKCECCQVSRQKSLSTMLALEESHAKY-CYFINPTYNDFN 66
I C +PI H G+ F C +C C +++ L LE S+A C FI TY+ +
Sbjct 12 IGCINPI--HIGNRYF--ACGRCSACLLAKSNKNRYNLTLELSNATTKCCFIMLTYDKEH 67
Query 67 IPAVRVPDDADFGSIA-EFEIITPRLKRDKYFEPYIVDYDVDLEKSIGQLCEQRDEYSRL 125
+P VR+ DF ++ + I P ++ +F + + S+ + YS
Sbjct 68 LPLVRI-SKHDFDAMYYKKPINKPEYEKRNFFCQLSYEKQLSKITSLSNRKVFKSAYSSQ 126
Query 126 --YSRSHKF-------VPHDVIYLL---HYPDIQRFIKRFRIYAKRKFNAS-CRYYVVGE 172
YS S F V D Y+L Y D+ F+KR R +R+ S R+ GE
Sbjct 127 SGYSMSTLFESGYNNSVHTDCYYMLPTLRYVDVSGFLKRLRTRVQREIGESNIRFAACGE 186
Query 173 YGTNSLRPHWHLLLFFDSDELAQDLERCY 201
YG RPH+H+++ S+ Q + R Y
Sbjct 187 YGPRGFRPHYHIIVICQSEAARQSVMRNY 215
>gi|490477382|ref|WP_004347759.1| hypothetical protein [Prevotella buccalis]
gi|281300711|gb|EFA93042.1| hypothetical protein HMPREF0650_1078 [Prevotella buccalis ATCC
35310]
Length=582
Score = 56.6 bits (135), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 51/187 (27%), Positives = 79/187 (42%), Gaps = 30/187 (16%)
Query 27 CRKCECCQVSRQKSLSTMLALEESHAKYCYFINPTYNDFNIPAVRVPDDADFGSIAEFEI 86
C KC C + +LS E KY F TY++ ++P V D++ E+
Sbjct 32 CDKCTACLNQKATTLSNRARAEIEQHKYSVFFTLTYDNEHLPKYEVFQDSN-------EV 84
Query 87 ITPRLKRDKYFEPYIVDYDVDLEKSIGQLC--EQRDEYSRLYS-RSHKFVP----HDVIY 139
I + P D + C + + Y LY F+P ++ IY
Sbjct 85 IQ--------YRPIGRLVDDSSSDMLSNSCPINKYNNYENLYQFDESTFIPPIENYEDIY 136
Query 140 ---LLHYPDIQRFIKRFR-----IYAKRKFNASCRYYVVGEYGTNSLRPHWHLLLFFDSD 191
++ DIQ F+KR R I K + RYY+ EYG + RPH+H +LFFDS
Sbjct 137 HFGVVCKKDIQNFLKRLRWRISKIPNITKDESKIRYYISSEYGPTTYRPHYHGILFFDSK 196
Query 192 ELAQDLE 198
++ ++
Sbjct 197 KILDKIK 203
>gi|575094436|emb|CDL65809.1| unnamed protein product [uncultured bacterium]
Length=340
Score = 55.5 bits (132), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 46/175 (26%), Positives = 70/175 (40%), Gaps = 47/175 (27%)
Query 16 IHRGSSIFLYPCRKCECCQVSRQKSLSTMLALE-ESHAKYCYFINPTYNDFNIPAVRVPD 74
IHR + L PC KC C++ ++ +T L LE ++ +F+ TY D IP
Sbjct 41 IHR-QDVMLIPCGKCIGCRIRAKQDWATRLELEARAYKGRAWFVTLTYRDDTIPL----- 94
Query 75 DADFGSIAEFEIITPRLKRDKYFEPYIVDYDVDLEKSIGQLCEQRDEYSRLYSRSHKFVP 134
L ++ G+L E ++SR VP
Sbjct 95 ---------------------------------LIRNTGELIEGG---VSMWSRGAD-VP 117
Query 135 HDVIYLLHYPDIQRFIKRFRIYAKRK--FNASCRYYVVGEYGTNSLRPHWHLLLF 187
I L+ D+ +F KR R Y + RY+ GEYG + RPH+H ++F
Sbjct 118 EQ-INTLNMDDVTKFWKRLRKYQTTEPDMGKELRYFYAGEYGEQTGRPHYHAIIF 171
Lambda K H a alpha
0.326 0.140 0.455 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 3643379990820