bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-4_CDS_annotation_glimmer3.pl_2_3
Length=638
Score E
Sequences producing significant alignments: (Bits) Value
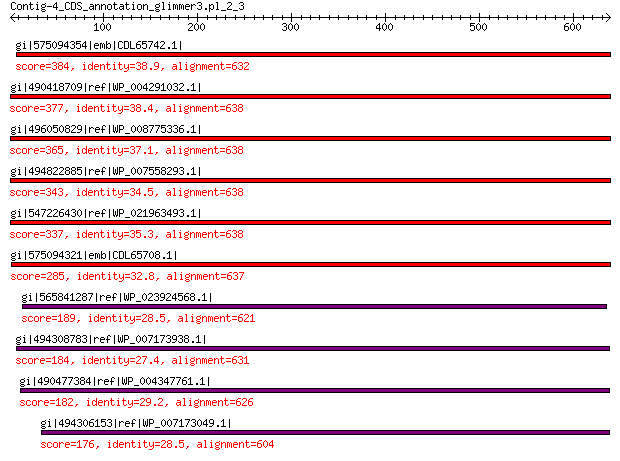
gi|575094354|emb|CDL65742.1| unnamed protein product 384 7e-121
gi|490418709|ref|WP_004291032.1| hypothetical protein 377 9e-119
gi|496050829|ref|WP_008775336.1| hypothetical protein 365 6e-114
gi|494822885|ref|WP_007558293.1| hypothetical protein 343 4e-105
gi|547226430|ref|WP_021963493.1| putative uncharacterized protein 337 3e-103
gi|575094321|emb|CDL65708.1| unnamed protein product 285 5e-83
gi|565841287|ref|WP_023924568.1| hypothetical protein 189 3e-48
gi|494308783|ref|WP_007173938.1| hypothetical protein 184 4e-47
gi|490477384|ref|WP_004347761.1| capsid protein 182 2e-46
gi|494306153|ref|WP_007173049.1| hypothetical protein 176 1e-44
>gi|575094354|emb|CDL65742.1| unnamed protein product [uncultured bacterium]
Length=615
Score = 384 bits (986), Expect = 7e-121, Method: Compositional matrix adjust.
Identities = 246/669 (37%), Positives = 357/669 (53%), Gaps = 93/669 (14%)
Query 7 LKELQNRPHKSGHDISAKNCFTAKVGELLPVWTDFAIPNCTYRINLEYFTRTRPVQTSAY 66
+ +++NRP ++G D+S K FTAK GELLPV T +P ++ INL FTRT+P+ TSA+
Sbjct 3 MADIKNRPSRNGFDLSFKKNFTAKAGELLPVMTKVVLPGDSFNINLRSFTRTQPLNTSAF 62
Query 67 TRIREYFDFFAVPCDLIWKSFDSAVIQMGEKAPIQSKDLLT-NLTVKGDLPYCSLSDLSF 125
R+REY+DF+ VP + +W FDS + QM S L N + G +PY + ++
Sbjct 63 ARMREYYDFYFVPFEQMWNKFDSCITQMNANVQHASGPTLDDNTPLSGRMPYFTSEQIAD 122
Query 126 ALKFASGNPASLGNKVSVSPGFGNIFGYNRGDVNHKLLSMLDYGNVVDKSANWIGTNANR 185
L + A+ K N FG+NR + KLL L YG+ N + N
Sbjct 123 YLN----DQATAARK--------NPFGFNRSTLTCKLLQYLGYGDY-----NSFDSETNT 165
Query 186 WWNRRVATADSAVYSQKYNVNLAVNLFFLATYQKIYQDFFRWSQWENADPTSYNFDWYRG 245
W +A +Y NL ++ F L YQKIY DF+R++QWE +P+++N D+ +G
Sbjct 166 W------SAKPLLY------NLELSPFPLLAYQKIYSDFYRYTQWEKTNPSTFNLDYIKG 213
Query 246 SGNLFGSSDLAAAIPASSDYWKRDNLFSLRYANWNKDKFMGILPNSQFGDLAVV------ 299
+ +L DL +P+ + N F +RY N+ KD F G+LP +Q+G +VV
Sbjct 214 TSDL--QMDLTG-LPSDDN-----NFFDIRYCNYQKDMFHGVLPVAQYGSASVVPINGQL 265
Query 300 ------DLGTISASGSKIP---------VGAYDGVNDSGTFKQFQTSSESYN--GSSDKT 342
D G I + + P VG GV++ + F S + N S+D +
Sbjct 266 NVISNGDSGPIFKTSTPDPGTPGTSYVTVGGNIGVDN----RSFGVSGSTLNVGKSADPS 321
Query 343 TQIYPRNPDSISVRADSNLWAVLGESSDLKLK------FSVLALRQAEALQKWKEITQSV 396
+P N + S+ LW E+ +L ++ +LALRQAE LQKWKE++ S
Sbjct 322 GYGFPSNASTRSL-----LW----ENPNLIIENNQGFYVPILALRQAEFLQKWKEVSVSG 372
Query 397 DTNYRDQIKAHFGVSVPQSEAHMAKYIGGVARNLDISEVVNNFLPGPGNDSSQAYIYGKG 456
+ +Y+ QI+ H+G+ V +H A+Y+GG A +LDI+EV+NN + G + A I GKG
Sbjct 373 EEDYKSQIEKHWGIKVSDFLSHQARYLGGCATSLDINEVINNNITG----DNAADIAGKG 428
Query 457 VGSGQGSMTFNTGSGYYILMCIYHAVPLLDYSISGPDGQNLVTSVEDLPIPEFDNIGMES 516
+G GS+ F + Y I+MCIYH +P++DY SG D + PIPE D IGMES
Sbjct 429 TFTGNGSIRFESKGEYGIIMCIYHVLPIVDYVGSGVDHSCTLVDATSFPIPELDQIGMES 488
Query 517 VPAVELMNSSLYSNVNSADKILGYSPRYYNWKTKIDRIHGAFTTTLKDWVAPIDDSFLYS 576
VP V MN S+ SAD LGY+PRY +WKT +DR G F +L+ W P+ D L S
Sbjct 489 VPLVRAMNPVKESDTPSADTFLGYAPRYIDWKTSVDRSVGDFADSLRTWCLPVGDKELTS 548
Query 577 LFGDSLS-------TYKGVTWPFFKVNPNTLDDLFAVKVDSTWETDQLLVNCNVGCYVTR 629
+SL+ + FFKVNP+ +D LFAV DST +TD+ L + V R
Sbjct 549 --ANSLNFPSNPNVEPDSIAAGFFKVNPSIVDPLFAVVADSTVKTDEFLCSSFFDVKVVR 606
Query 630 PLSADGVPY 638
L +G+PY
Sbjct 607 NLDVNGLPY 615
>gi|490418709|ref|WP_004291032.1| hypothetical protein [Bacteroides eggerthii]
gi|217986636|gb|EEC52970.1| putative capsid protein (F protein) [Bacteroides eggerthii DSM
20697]
Length=578
Score = 377 bits (968), Expect = 9e-119, Method: Compositional matrix adjust.
Identities = 245/661 (37%), Positives = 342/661 (52%), Gaps = 106/661 (16%)
Query 1 MAHFTGLKELQNRPHKSGHDISAKNCFTAKVGELLPVWTDFAIPNCTYRINLEYFTRTRP 60
MA+ LK ++N+P ++G D+S K FTAK GELLPV +P T++INL+ FTRT+P
Sbjct 1 MANIMSLKSIRNKPSRNGFDLSFKKNFTAKAGELLPVMVKEVLPGDTFKINLKAFTRTQP 60
Query 61 VQTSAYTRIREYFDFFAVPCDLIWKSFDSAVIQMGEKAP-IQSKDLLTNLTVKGDLPYCS 119
V T+A+ RIREY+DFF VP DL+W ++ + QM + S D N + G++PY +
Sbjct 61 VNTAAFARIREYYDFFFVPYDLLWNKANTVLTQMYDNPQHAVSIDPTRNFVLSGEMPYMT 120
Query 120 LSDLSFALKFASGNPASLGNKVSVSPGFG----NIFGYNRGDVNHKLLSMLDYGNVVDKS 175
+ AS N +S + N FGYNR + KLL L YGN
Sbjct 121 SEAI-----------ASYINALSTASALADYKSNYFGYNRSKSSVKLLEYLGYGNY---- 165
Query 176 ANWIGTNANRWWNRRVATADSAVYSQKYNVNLAVNLFFLATYQKIYQDFFRWSQWENADP 235
+++ + WN A NL N+F L YQKIY DF+R SQWE P
Sbjct 166 ESFLTDD----WNTAPLMA-----------NLNHNIFGLLAYQKIYSDFYRDSQWERVSP 210
Query 236 TSYNFDWYRGSGNLFGSSDLAAAIPASSDYWKRDNLFSLRYANWNKDKFMGILPNSQFGD 295
+++N D+ GS S +L A S+++++ N F LRY NW KD F G+LP+ Q+G+
Sbjct 211 STFNVDYLDGS-----SMNLDNAY--STEFYQNYNFFDLRYCNWQKDLFHGVLPHQQYGE 263
Query 296 LAVVDLGTISASGSKIPVGAYDGVNDSGTFKQFQTSSESYNGSSDKTTQIYPRNPDSISV 355
AV + T +G T F T S P + S
Sbjct 264 TAVASI-TPDVTGKL-------------TLSNFSTVGTS---------------PTTASG 294
Query 356 RADSNLWAVLGESSDLKLKFSVLALRQAEALQKWKEITQSVDTNYRDQIKAHFGVSVPQS 415
A NL A D S+L LRQAE LQKWKEITQS + +Y+DQ++ H+GVSV
Sbjct 295 TATKNLPAF-----DTVGDLSILVLRQAEFLQKWKEITQSGNKDYKDQLEKHWGVSVGDG 349
Query 416 EAHMAKYIGGVARNLDISEVVNNFLPGPGNDSSQAYIYGKGVGSGQGSMTFNTGSGYYIL 475
+ + Y+GGV+ ++DI+EV+N + G S+ A I GKGVG G + FN+ Y ++
Sbjct 350 FSELCTYLGGVSSSIDINEVINTNITG----SAAADIAGKGVGVANGEINFNSNGRYGLI 405
Query 476 MCIYHAVPLLDYSISGPDGQNLVTSVEDLPIPEFDNIGMESVPAVELMNSSLYSNVNSAD 535
MCIYH +PLLDY+ D L + D IPEFD +GM+S+P V+LMN L S N++
Sbjct 406 MCIYHCLPLLDYTTDMLDPAFLKVNSTDYAIPEFDRVGMQSMPLVQLMN-PLRSFANASG 464
Query 536 KILGYSPRYYNWKTKIDRIHGAFTTTLKDWV------------------APIDDSFLYSL 577
+LGY PRY ++KT +D+ G F TL WV PI+ S
Sbjct 465 LVLGYVPRYIDYKTSVDQSVGGFKRTLNSWVISYGNISVLKQVTLPNDAPPIEPS----- 519
Query 578 FGDSLSTYKGVTWPFFKVNPNTLDDLFAVKVDSTWETDQLLVNCNVGCYVTRPLSADGVP 637
+ + + + + FFKVNP+ LD +FAV+ TDQ L + R L DG+P
Sbjct 520 --EPVPSVAPMNFTFFKVNPDCLDPIFAVQAGDDTNTDQFLCSSFFDIKAVRNLDTDGLP 577
Query 638 Y 638
Y
Sbjct 578 Y 578
>gi|496050829|ref|WP_008775336.1| hypothetical protein [Bacteroides sp. 2_2_4]
gi|229448893|gb|EEO54684.1| putative capsid protein (F protein) [Bacteroides sp. 2_2_4]
Length=580
Score = 365 bits (936), Expect = 6e-114, Method: Compositional matrix adjust.
Identities = 237/644 (37%), Positives = 344/644 (53%), Gaps = 70/644 (11%)
Query 1 MAHFTGLKELQNRPHKSGHDISAKNCFTAKVGELLPVWTDFAIPNCTYRINLEYFTRTRP 60
MA+ LK L+N+ ++G D+S+K FTAK GELLPV +P + I+L+ FTRT+P
Sbjct 1 MANIMSLKSLRNKTSRNGFDLSSKRNFTAKPGELLPVKCWEVLPGDKWSIDLKSFTRTQP 60
Query 61 VQTSAYTRIREYFDFFAVPCDLIWKSFDSAVIQMGEKAPIQSKDLL--TNLTVKGDLPYC 118
+ T+A+ R+REY+DF+ VP +L+W ++ + QM + P + + N + G +P
Sbjct 61 LNTAAFARMREYYDFYFVPYNLLWNKANTVLTQMYDN-PQHATSYIPSANQALAGVMPNV 119
Query 119 SLSDLSFALKFASGNPASLGNKVSVSPGFGNIFGYNRGDVNHKLLSMLDYGNVVDKSANW 178
+ ++ L + + + + N FGY+R KLL L YGN
Sbjct 120 TCKGIADYLNLVAPDVTTTNSYEK------NYFGYSRSLGTAKLLEYLGYGNFYT----- 168
Query 179 IGTNANRWWNRRVATADSAVYSQKYNVNLAVNLFFLATYQKIYQDFFRWSQWENADPTSY 238
T+ N W + ++ NL +N++ + YQKIY D R SQWE P+ +
Sbjct 169 YATSKNNTWTKSPLSS-----------NLQLNIYGVLAYQKIYADHIRDSQWEKVSPSCF 217
Query 239 NFDWYRGSGNLFGSSDLAAAIPASSDYWKRDNLFSLRYANWNKDKFMGILPNSQFGDLAV 298
N D+ G+ + + D + ++ N+F LRY NW KD F G+LP Q+GD A
Sbjct 218 NVDYLSGTVDSAMTIDSMITGQGFAPFY---NMFDLRYCNWQKDLFHGVLPRQQYGDTAA 274
Query 299 VDLGTISASGSKIPVGAYDGVNDSGTFKQFQTSSESYNGSSDKTTQIYPRNPDSISVRAD 358
V++ + ++ V DG + GS +T +
Sbjct 275 VNVNLSNVLSAQYMVQTPDG--------------DPVGGSPFSSTGV------------- 307
Query 359 SNLWAVLGESSDLKLKFSVLALRQAEALQKWKEITQSVDTNYRDQIKAHFGVSVPQSEAH 418
NL V G + F+VLALRQAE LQKWKEITQS + +Y+DQI+ H+ VSV ++ +
Sbjct 308 -NLQTVNGSGT-----FTVLALRQAEFLQKWKEITQSGNKDYKDQIEKHWNVSVGEAYSE 361
Query 419 MAKYIGGVARNLDISEVVNNFLPGPGNDSSQAYIYGKGVGSGQGSMTFNTGSGYYILMCI 478
M+ Y+GG +LDI+EVVNN + G S+ A I GKGV G G ++F+ G Y ++MCI
Sbjct 362 MSLYLGGTTASLDINEVVNNNITG----SNAADIAGKGVVVGNGRISFDAGERYGLIMCI 417
Query 479 YHAVPLLDYSISGPDGQNLVTSVEDLPIPEFDNIGMESVPAVELMNSSLYSNVNSADKIL 538
YH++PLLDY+ + + D IPEFD +GMESVP V LMN L S+ N IL
Sbjct 418 YHSLPLLDYTTDLVNPAFTKINSTDFAIPEFDRVGMESVPLVSLMN-PLQSSYNVGSSIL 476
Query 539 GYSPRYYNWKTKIDRIHGAFTTTLKDWVAPIDD-SFLYSL-FGDSLSTYKG--VTWPFFK 594
GY+PRY ++KT +D GAF TTLK WV D+ S + L + D + G V + FK
Sbjct 477 GYAPRYISYKTDVDSSVGAFKTTLKSWVMSYDNQSVINQLNYQDDPNNSPGTLVNYTNFK 536
Query 595 VNPNTLDDLFAVKVDSTWETDQLLVNCNVGCYVTRPLSADGVPY 638
VNPN +D LFAV ++ +TDQ L + V R L DG+PY
Sbjct 537 VNPNCVDPLFAVAASNSIDTDQFLCSSFFDVKVVRNLDTDGLPY 580
>gi|494822885|ref|WP_007558293.1| hypothetical protein [Bacteroides plebeius]
gi|198272099|gb|EDY96368.1| putative capsid protein (F protein) [Bacteroides plebeius DSM
17135]
Length=613
Score = 343 bits (879), Expect = 4e-105, Method: Compositional matrix adjust.
Identities = 220/651 (34%), Positives = 335/651 (51%), Gaps = 58/651 (9%)
Query 1 MAHFTGLKELQNRPHKSGHDISAKNCFTAKVGELLPVWTDFAIPNCTYRINLEYFTRTRP 60
MA+ +K ++N+P ++G+D++ K FTAK G L+PVW +P ++ F RT+P
Sbjct 8 MANIMSMKSVRNKPTRAGYDLTQKINFTAKAGSLIPVWWTPVLPFDDLNATVKSFVRTQP 67
Query 61 VQTSAYTRIREYFDFFAVPCDLIWKSFDSAVIQMGEKAPIQSKDLLT-NLTVKGDLPYCS 119
+ T+A+ R+R YFDF+ VP +W F +A+ QM S +L N+ + +LPY +
Sbjct 68 LNTAAFARMRGYFDFYFVPFRQMWNKFPTAITQMRTNLLHASGPVLADNVPLSDELPYFT 127
Query 120 LSDLS-FALKFASGNPASLGNKVSVSPGFGNIFGYNRGDVNHKLLSMLDYGNVVDKSANW 178
++ + + A N FGY R + +L L YG+
Sbjct 128 AEQVADYIVSLADSK---------------NQFGYYRAWLVCIILEYLGYGDFYPYIVEA 172
Query 179 IGTNANRWWNRRVATADSAVYSQKYNVNLAVNLFFLATYQKIYQDFFRWSQWENADPTSY 238
G W R + NL + F L YQKIY DF R++QWE ++P+++
Sbjct 173 AGGEGATWATRPMLN------------NLKFSPFPLFAYQKIYADFNRYTQWERSNPSTF 220
Query 239 NFDWYRGSGNLFGSSDLAAAIPASSDYWKRDNLFSLRYANWNKDKFMGILPNSQFGDLAV 298
N D+ GS + S L + D + NLF +RY+NW +D G +P +Q+G+ +
Sbjct 221 NIDYISGSAD---SLQLDFTVEGFKDSF---NLFDMRYSNWQRDLLHGTIPQAQYGEASA 274
Query 299 VDL-GTISASGSKIPVGAYDGVNDSGTFKQFQTSSESYNGSSDKTTQIYPRNPDSISVRA 357
V + G++ P A+ D F + + +G T + +S +R
Sbjct 275 VPVSGSMQVVEGPTP-PAFTTGQDGVAFLNGNVTIQGSSGYLQAQTSV----GESRILRF 329
Query 358 DSNLWAVLGESSDLKLKFSVLALRQAEALQKWKEITQSVDTNYRDQIKAHFGVSVPQSEA 417
++ ++ E D S+LALR+AEA QKWKE+ + + +Y QI+AH+G SV ++ +
Sbjct 330 NNTNSGLIVE-GDSSFGVSILALRRAEAAQKWKEVALASEEDYPSQIEAHWGQSVNKAYS 388
Query 418 HMAKYIGGVARNLDISEVVNNFLPGPGNDSSQAYIYGKGVGSGQGSMTFNTGSGYYILMC 477
M +++G + +L I+EVVNN + G + A I GKG SG GS+ FN G Y I+MC
Sbjct 389 DMCQWLGSINIDLSINEVVNNNITG----ENAADIAGKGTMSGNGSINFNVGGQYGIVMC 444
Query 478 IYHAVPLLDYSISGPDGQNLVTSVEDLPIPEFDNIGMESVPAVELMNSSLYSNVN---SA 534
++H +P LDY S P +T+V D PIPEFD IGME VP + +N + + S
Sbjct 445 VFHVLPQLDYITSAPHFGTTLTNVLDFPIPEFDKIGMEQVPVIRGLNPVKPKDGDFKVSP 504
Query 535 DKILGYSPRYYNWKTKIDRIHGAFTTTLKDWVAPIDDSFLYSLFGDSLS-------TYKG 587
+ GY+P+YYNWKT +D+ G F +LK W+ P DD L L DS+
Sbjct 505 NLYFGYAPQYYNWKTTLDKSMGEFRRSLKTWIIPFDDEAL--LAADSVDFPDNPNVEADS 562
Query 588 VTWPFFKVNPNTLDDLFAVKVDSTWETDQLLVNCNVGCYVTRPLSADGVPY 638
V FFKV+P+ LD+LFAVK +S TDQ L + V R L +G+PY
Sbjct 563 VKAGFFKVSPSVLDNLFAVKANSDLNTDQFLCSTLFDVNVVRSLDPNGLPY 613
>gi|547226430|ref|WP_021963493.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
gi|524103382|emb|CCY83994.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
Length=573
Score = 337 bits (863), Expect = 3e-103, Method: Compositional matrix adjust.
Identities = 225/650 (35%), Positives = 341/650 (52%), Gaps = 89/650 (14%)
Query 1 MAHFTGLKELQNRPHKSGHDISAKNCFTAKVGELLPVWTDFAIPNCTYRINLEYFTRTRP 60
M+ L L+N ++G D+S KN FTAKVGELLP+ P + I + FTRT+P
Sbjct 1 MSSVMSLTALKNSVKRNGFDLSFKNAFTAKVGELLPIMCKEVYPGDKFNIRGQAFTRTQP 60
Query 61 VQTSAYTRIREYFDFFAVPCDLIWKSFDSAVIQMGEKAPIQSKDLLTNLTVKGDLPYCSL 120
V ++AY+R+REY+DF+ VP L+W + M + P + DL++++ + P+ +
Sbjct 61 VNSAAYSRLREYYDFYFVPYRLLWNMAPTFFTNMPD--PHHAADLVSSVNLSQRHPWFTF 118
Query 121 SDLSFALKFASGNPASLGNKVSVSPGF----GNIFGYNRGDVNHKLLSMLDYGNVVDKSA 176
D+ LGN S+S + N FG++R +++ KLL+ L+YG D +
Sbjct 119 FDIM----------EYLGNLNSLSGAYEKYQKNFFGFSRVELSVKLLNYLNYGFGKDYES 168
Query 177 NWIGTNANRWWNRRVATADSAVYSQKYNVNLAVNLFFLATYQKIYQDFFRWSQWENADPT 236
+ ++++ ++ ++ F L YQKI +D+FR QW++A P
Sbjct 169 VKVPSDSD---------------------DIVLSPFPLLAYQKICEDYFRDDQWQSAAPY 207
Query 237 SYNFDWYRGSGNLFGSSDLAAAIPASS---DYWKRDNLFSLRYANWNKDKFMGILPNSQF 293
YN D+ G + F IP SS D +K +F L Y N+ KD F G+LP +Q+
Sbjct 208 RYNLDYLYGKSSGF-------HIPMSSFTNDAFKNPTMFDLNYCNFQKDYFTGMLPRAQY 260
Query 294 GDLAVVDLGTISASGSKIPVGAYDGVNDSGTFKQFQTSSESYNGSSDKTTQIYPRNPDSI 353
GD++V P+ + DS + +S G++ + +
Sbjct 261 GDVSVAS-----------PIFGDLDIGDSSSLT---FASAPQQGANTIQSGV-------- 298
Query 354 SVRADSNLWAVLGESSDLKLKFSVLALRQAEALQKWKEITQSVDTNYRDQIKAHFGVSVP 413
V+ +S+ SVLALRQAE LQKW+EI QS +Y+ Q++ HF VS
Sbjct 299 ---------LVVNNNSNTTAGLSVLALRQAECLQKWREIAQSGKMDYQTQMQKHFNVSPS 349
Query 414 QSEAHMAKYIGGVARNLDISEVVNNFLPGPGNDSSQAYIYGKGVGSGQGSMTFNTGSGYY 473
+ + KY+GG NLDISEVVN L G +QA I GKG G+ G+ S +
Sbjct 350 ATLSGHCKYLGGWTSNLDISEVVNTNLTG----DNQADIQGKGTGTLNGNKVDFESSEHG 405
Query 474 ILMCIYHAVPLLDYSISGPDGQNLVTSVEDLPIPEFDNIGMESVPAVELMN--SSLYSNV 531
I+MCIYH +PLLD+SI+ QN T+ D IPEFD++GM+ + E++ L S+
Sbjct 406 IIMCIYHCLPLLDWSINRIARQNFKTTFTDYAIPEFDSVGMQQLYPSEMIFGLEDLPSDP 465
Query 532 NSADKILGYSPRYYNWKTKIDRIHGAFTTTLKDWVAPIDDSFLYSL---FGDSLSTYKGV 588
+S + +GY PRY + KT ID IHG+F TL WV+P+ DS++ + D+ + +
Sbjct 466 SSIN--MGYVPRYADLKTSIDEIHGSFIDTLVSWVSPLTDSYISAYRQACKDAGFSDITM 523
Query 589 TWPFFKVNPNTLDDLFAVKVDSTWETDQLLVNCNVGCYVTRPLSADGVPY 638
T+ FFKVNP+ +D++F VK DST TDQLL+N R +G+PY
Sbjct 524 TYNFFKVNPHIVDNIFGVKADSTINTDQLLINSYFDIKAVRNFDYNGLPY 573
>gi|575094321|emb|CDL65708.1| unnamed protein product [uncultured bacterium]
Length=642
Score = 285 bits (730), Expect = 5e-83, Method: Compositional matrix adjust.
Identities = 209/666 (31%), Positives = 334/666 (50%), Gaps = 59/666 (9%)
Query 2 AHFTGLKELQNRPHKSGHDISAKNCFTAKVGELLPVWTDFAIPNCTYRINLEYFTRTRPV 61
++ GL L+N+P ++ D+S +N FTAKVGELLP + P + +++ YFTRT P+
Sbjct 5 SNIMGLHGLKNKPSRNSFDLSHRNMFTAKVGELLPCFVQELNPGDSVKVSSSYFTRTAPL 64
Query 62 QTSAYTRIREYFDFFAVPCDLIWKSFDSAVIQM------GEKAPIQSKDLLTNLTVKGDL 115
Q++A+TR+RE +F VP +WK FDS V+ M G+ + I S L+ N V +
Sbjct 65 QSNAFTRLRENVQYFFVPYSALWKYFDSQVLNMTKNANGGDISRIASS-LVGNQKVTTQM 123
Query 116 PYCSLSDL-SFALKFASGNPASLGNKVSVSPGFGNIFGYNRGDVNH----KLLSMLDYGN 170
P + L ++ LKF N +++G+ SV P F NRG H KLL +L YGN
Sbjct 124 PCVNYKTLHAYLLKFI--NRSTVGSDGSVGPEF------NRGCYRHAESAKLLQLLGYGN 175
Query 171 VVDKSANWIGTNANRWWNRRVATADSAVYSQKYNVNLAVNLFFLATYQKIYQDFFRWSQW 230
++ AN+ + N + + YN + +++F L Y KI D + + QW
Sbjct 176 FPEQFANF------KVNNDKHNQSGQNFKDVTYNNSPYLSIFRLLAYHKICNDHYLYRQW 229
Query 231 ENADPTSYNFDWYR-GSGNLFGSSDLAAAIPASSDYWKRDNLFSLRYANWNKDKFMGILP 289
+ + + N D+ S +L D +IP S ++ NL +R++N D F G+LP
Sbjct 230 QPYNASLCNVDYLTPNSSSLLSIDDALLSIPDDSIKAEKLNLLDMRFSNLPLDYFTGVLP 289
Query 290 NSQFGDLAVVDLGTISASGSKIPVGAYDG-----VNDSGTFKQFQTSSESYNGSSDKTTQ 344
SQFG +VV+L +ASGS + G +G ++ Q + S NG+
Sbjct 290 TSQFGSESVVNLNLGNASGSAVLNGTTSKDSGRWRTTTGEWEMEQRVASSANGN------ 343
Query 345 IYPRNPDSISVRADSNLWAVLGESSDLKLKFSVLALRQAEALQKWKEITQSVDTNYRDQI 404
+ N + + D + ++ L S++ALR A A QK+KEI + D +++ Q+
Sbjct 344 LKLDNSNGTFISHDHTFSGNVAINTSLSGNLSIIALRNALAAQKYKEIQLANDVDFQSQV 403
Query 405 KAHFGVSVPQSEAHMAKYIGGVARNLDISEVVNNFLPGPGNDSSQAYIYGKGVGSGQGSM 464
+AHFG+ P + + +IGG + ++I+E +N L G ++A G+G S+
Sbjct 404 EAHFGIK-PDEKNENSLFIGGSSSMININEQINQNLSG----DNKATYGAAPQGNGSASI 458
Query 465 TFNTGSGYYILMCIYHAVPLLDYSISGPDGQNLVTSVEDLPIPEFDNIGMESVPAVELMN 524
F T Y +++ IY P+LD++ G D T D IPE D+IGM+ E+
Sbjct 459 KF-TAKTYGVVIGIYRCTPVLDFAHLGIDRTLFKTDASDFVIPEMDSIGMQQTFRCEVAA 517
Query 525 SSLYSNVNSADKI-----------LGYSPRYYNWKTKIDRIHGAFTTTLKDWVAPIDDSF 573
+ Y++ A ++ GY+PRY +KT DR +GAF +LK WV I+
Sbjct 518 PAPYNDEFKAFRVGDGSSPDMSETYGYAPRYSEFKTSYDRYNGAFCHSLKSWVTGIN--- 574
Query 574 LYSLFGDSLSTYKGVTWP-FFKVNPNTLDDLFAVKVDSTWETDQLLVNCNVGCYVTRPLS 632
++ + +T+ G+ P F P+ + +LF V + + DQL V CY TR LS
Sbjct 575 FDAIQNNVWNTWAGINAPNMFACRPDIVKNLFLVSSTNNSDDDQLYVGMVNMCYATRNLS 634
Query 633 ADGVPY 638
G+PY
Sbjct 635 RYGLPY 640
>gi|565841287|ref|WP_023924568.1| hypothetical protein [Prevotella nigrescens]
gi|564729907|gb|ETD29851.1| hypothetical protein HMPREF1173_00033 [Prevotella nigrescens
CC14M]
Length=656
Score = 189 bits (480), Expect = 3e-48, Method: Compositional matrix adjust.
Identities = 177/699 (25%), Positives = 301/699 (43%), Gaps = 134/699 (19%)
Query 14 PH----KSGHDISAKNCFTAKVGELLPVWTDFAIPNCTYRINLEYFTRTRPVQTSAYTRI 69
PH ++G+D+S++ F+A G LLP+ T A P +RI+++ R +P+ T+A+ R
Sbjct 9 PHPNLNRNGYDLSSRRIFSAPAGALLPIATWEANPGEKFRISVQDLVRAQPLNTAAFARC 68
Query 70 REYFDFFAVPCDLIWKSFDSAVIQMGEKAPIQSK-DLLTNLT-VKGDLPYCSLSDLSFAL 127
+EY+ FF VP +W+ D + E SK D T+ V +P +L D+ +
Sbjct 69 KEYYHFFFVPYKSLWQHSDRFFTGVTEGDSAFSKPDGKTDFNFVPKTVPMFNLKDV---V 125
Query 128 KFASGNPASLGNKVSVSPGFGNIFGYNRGDVNHKLLSMLDYG------------NVVDKS 175
+ A L N ++ G G RG +L + G +++K
Sbjct 126 SLLHASTADLFNSENLGISKGKGIGVTRGSYQSSILDDIHSGTKTNPIDRKKLDELIEKM 185
Query 176 ANWIGTNANRWWNR----RVATADSAVYSQKYNV-------------------------- 205
+ + +++R ++++ D+ Y K+
Sbjct 186 RLALPADLKPYYDRFVVSKLSSKDALGYLYKFGAFRLLHFLGYGVDNNGFIVDFNASYAA 245
Query 206 ------------------NLAVNLFFLATYQKIYQDFFRWSQWENADPTSYNFDWYRGSG 247
++ N+F L YQ+IY DF+R WE A P +N DW +
Sbjct 246 GTGEIVKNVLAKKTYKLPDIKANVFRLLAYQRIYNDFYRNDLWEAAQPDVFNVDWCCNNN 305
Query 248 NLFGSSDLAAAIPASSDYWKRDNLFSLRYANWNKDKFMGILPNSQFGDLAVVDLGTISAS 307
+L S +L + LRY +W+KD P + + D + +L
Sbjct 306 SLDISDELVY------------KMCQLRYRHWSKDWVTSAYPTASY-DKGIFELP----- 347
Query 308 GSKIPVGAYDGVNDSGTFKQFQTSSESYNGSSDKTTQIYPRNPDSISVRADSNLWAVLGE 367
D +N + F + + N ++ +Q+ ++ D+ S+ +++ +
Sbjct 348 ---------DYINGNTGFATTEVKRDVVN---NRGSQLEIKSMDAGSLGSNNISYI---S 392
Query 368 SSDLKLKFSVLALRQAEALQKWKEITQSVD-TNYRDQIKAHFGVSVPQSEAHMAKYIGGV 426
+D++ F AL+K E T++ + +Y +QI AHFG VP+S + A +IGG
Sbjct 393 PNDIRAMF---------ALEKMLERTRAANGLDYSNQIAAHFGFKVPESRKNCASFIGGF 443
Query 427 ARNLDISEVVNNFLPGPGNDSSQAYIYGKGVGSGQGSMTFNTGSGYY------ILMCIYH 480
+ ISEVV +S + G+ G G G+M N+G Y ++MCIY
Sbjct 444 DNQISISEVVTTSNGSVDGTASTGSVVGQVFGKGIGAM--NSGHISYDVKEHGLIMCIYS 501
Query 481 AVPLLDYSISGPDGQNLVTSVEDLPIPEFDNIGMESVPAVEL---MNSSLYSNVNSADKI 537
P +DY D N S ED PEF+N+GM+ V +L +NS+ + + + +
Sbjct 502 IAPQVDYDARELDPFNRKFSREDYFQPEFENLGMQPVIQSDLCLCINSAKSDSSDQHNNV 561
Query 538 LGYSPRYYNWKTKIDRIHGAFTT--TLKDWVAPIDDSFLYSLFGDSLSTYKGVTWPFFKV 595
LGYS RY +KT D I G F + +L W P ++ Y+ + ++ P V
Sbjct 562 LGYSARYLEYKTARDIIFGEFMSGGSLSAWATPKNN---YTF------EFGKLSLPDLLV 612
Query 596 NPNTLDDLFAVKVDSTWETDQLLVNCNVGCYVTRPLSAD 634
+P L+ +FAVK + + TDQ LVN RP+ +
Sbjct 613 DPKVLEPIFAVKYNGSMSTDQFLVNSYFDVKAIRPMQVN 651
>gi|494308783|ref|WP_007173938.1| hypothetical protein [Prevotella bergensis]
gi|270333035|gb|EFA43821.1| putative capsid protein (F protein) [Prevotella bergensis DSM
17361]
Length=553
Score = 184 bits (468), Expect = 4e-47, Method: Compositional matrix adjust.
Identities = 173/646 (27%), Positives = 272/646 (42%), Gaps = 117/646 (18%)
Query 7 LKELQNRPHKSGHDISAKNCFTAKVGELLPVWTDFAIPNCTYRINLEYFTRTRPVQTSAY 66
+K + +++ D+S ++ FTA G LLPV IP+ IN + F RT P+ T+A+
Sbjct 8 IKATRPNRNRNAFDLSQRHLFTAHAGMLLPVLNLDLIPHDHVEINAQDFMRTLPMNTAAF 67
Query 67 TRIREYFDFFAVPCDLIWKSFDSAVIQMGEKAPIQSKDLLTNLTVKGDLPYCSLSDLSFA 126
+R ++FF VP +W FD + M + +K + T +PY ++ + +
Sbjct 68 ASMRGVYEFFFVPYHQLWAQFDQFITGMNDFHSSANKSIQGG-TSPLQVPYFNVDSVFNS 126
Query 127 LKFASGNPASLGNKVSVSPGFGNIFGYNRGDVNHKLLSMLDYGNVVDKSANWIGTNANRW 186
L + + + + +G +LL +L YG D N
Sbjct 127 LNTGKESGSGSTDDLQYKFKYGAF----------RLLDLLGYGRKFDSFGTAYPDN---- 172
Query 187 WNRRVATADSAVYSQKYNVNLAVNLFFLATYQKIYQDFFRWSQWENADPTSYNFDWYRGS 246
V K N++ ++F + Y KIYQD++R S +EN D S+NFD ++G
Sbjct 173 -----------VSGLKNNLDYNCSVFRILAYNKIYQDYYRNSNYENFDTDSFNFDKFKG- 220
Query 247 GNLFGSSDLAAAIPASSDYWKRDNLFSLRYANWNKDKFMGILPNSQFGDLAVVDLGTISA 306
G D A + A +LF LRY N D F + + F
Sbjct 221 ----GLVD--AKVVA--------DLFKLRYRNAQTDYFTNLRQSQLF------------- 253
Query 307 SGSKIPVGAYDGVNDSGTFKQFQTSSESYNGSSDKTTQIYPRNPDSISVRAD-SNLWAV- 364
F T+ E + I PR+ V++D SN V
Sbjct 254 --------------------SFTTAFEDVD-----NINIAPRD----YVKSDGSNFTRVN 284
Query 365 LGESSDL-KLKFSVLALRQAEALQKWKEITQSVDTNYRDQIKAHFGVSVPQSEAHMAKYI 423
G +D + FSV +LR A A+ K +T ++DQ++AH+GV +P S Y+
Sbjct 285 FGVDTDSSEGDFSVSSLRAAFAVDKLLSVTMRAGKTFQDQMRAHYGVEIPDSRDGRVNYL 344
Query 424 GGVARNLDISEVVN-------NFLPGPGNDSSQAYIYGKGVGSGQGSMTFNTGSGYYILM 476
GG ++ +S+V + P G + GKG GSG+G + F+ + +LM
Sbjct 345 GGFDSDMQVSDVTQTSGTTATEYKPEAG---YLGRVAGKGTGSGRGRIVFDAKE-HGVLM 400
Query 477 CIYHAVPLLDYSISGPDGQNLVTSVEDLPIPEFDNIGMESVPAVELMNSSLYSNVNSAD- 535
CIY VP + Y + D D PEF+N+GM+ +NSS S+ + D
Sbjct 401 CIYSLVPQIQYDCTRLDPMVDKLDRFDYFTPEFENLGMQP------LNSSYISSFCTTDP 454
Query 536 --KILGYSPRYYNWKTKIDRIHGAFTTT--LKDWVAPIDDSFLYSLFGDSLSTYKGVTWP 591
+LGY PRY +KT +D HG F + L W S F +T+ +
Sbjct 455 KNPVLGYQPRYSEYKTALDVNHGQFAQSDALSSWSV--------SRF-RRWTTFPQLEIA 505
Query 592 FFKVNPNTLDDLFAVKVDSTWETDQLLVNCNVGCYVTRPLSADGVP 637
FK++P L+ +F V + T D + CN +S DG+P
Sbjct 506 DFKIDPGCLNSIFPVDYNGTEANDCVYGGCNFNIVKVSDMSVDGMP 551
>gi|490477384|ref|WP_004347761.1| capsid protein [Prevotella buccalis]
gi|281300712|gb|EFA93043.1| putative capsid protein (F protein) [Prevotella buccalis ATCC
35310]
Length=552
Score = 182 bits (462), Expect = 2e-46, Method: Compositional matrix adjust.
Identities = 183/640 (29%), Positives = 271/640 (42%), Gaps = 117/640 (18%)
Query 12 NRPHKSGHDISAKNCFTAKVGELLPVWTDFAIPNCTYRINLEYFTRTRPVQTSAYTRIRE 71
NRP ++ D+S K+ FTA G LLPV T IP+ I F R P+ ++A+ +R
Sbjct 14 NRP-RNAFDLSQKHLFTAHAGMLLPVMTLDLIPHDHVSIQATDFMRCLPMNSAAFMSMRS 72
Query 72 YFDFFAVPCDLIWKSFDSAVIQMGEKAPIQSKDLLTNLTVKGDLPYCSLSDLSFALKFAS 131
++FF VP +W FD + M + + DL + K L S F +
Sbjct 73 VYEFFFVPYSQLWHPFDQFITGMNDYRSVLQSDLYKS---KSPLVIPSFKRKELYELFNA 129
Query 132 GNPASLGNKVSVSPGFGNIFGYNRGDVNHKLLSMLDYGNVVDKSANWIGTNANRWWNRRV 191
P N+ S P +IFG+ +LL +L YG V NA+
Sbjct 130 --PGGFLNQQSNQP---DIFGFKSRFNFLRLLDLLGYGVYV---------NAD------- 168
Query 192 ATADSAVYSQKYNVNLAVNLFFLATYQKIYQDFFRWSQWENADPTSYNFDWYRGSGNLFG 251
++ +S+ + +++F LA YQKIY DF+R + +E D +S++ D
Sbjct 169 GSSRIDAFSKLLDDTEKLSIFRLAAYQKIYSDFYRNTTYEAVDVSSFSLD---------N 219
Query 252 SSDLAAAIPASSDYWKRDNLFSLRYANWNKDKFMGILPNSQFGDLAVVDLGTISASGSKI 311
+D +AI A +KR +LRY N D F + P F DL
Sbjct 220 ITDSISAINA----FKR--FGTLRYRNAQLDYFTNLRPTPLF-DL--------------- 257
Query 312 PVGAYDGVNDSGTFKQFQTSSESYNGSSDKTTQIYPRNPDSISVRADSNLWAV-LGESSD 370
D+ + F YN P N DS+S+ +DSN AV SD
Sbjct 258 ---------DNPSLNSF------YNT---------PGNADSVSIDSDSN--AVNFQLDSD 291
Query 371 LKLKFSVLALRQAEALQKWKEITQSVDTNYRDQIKAHFGVSVPQSEAHMAKYIGGVARNL 430
L +V ++R A AL K ITQ Y +QIKAHFG V + YIGG N+
Sbjct 292 L---LTVQSIRNAFALDKLMRITQRAGKTYAEQIKAHFGFEVSEGRDGRVNYIGGFDSNI 348
Query 431 DISEVVNNFLPGPGNDSSQAY----------IYGKGVGSGQGSMTFNTGSGYYILMCIYH 480
+ +V + G Q + GK GSG G + F+ + ILMCIY
Sbjct 349 QVGDVTQ--MSGTTASPEQGVSIKHGGYLGRVTGKAQGSGSGHIEFDAHE-HGILMCIYS 405
Query 481 AVPLLDYSISGPDGQNLVTSVEDLPIPEFDNIGMESVPAVELMNSSLYSNVNS-ADKILG 539
VP + Y + D S D +PEF+++GM+ + + S++ + +K G
Sbjct 406 LVPDMQYDATRIDPFVTKLSRGDFFMPEFEDLGMQP------LQTRYISDIRTQTEKFKG 459
Query 540 YSPRYYNWKTKIDRIHGAFTT--TLKDWVAPIDDSFLYSLFGDSLSTYKGVTWPFFKVNP 597
+ PRY +KT +D HG F L W + G++L T+ + K+NP
Sbjct 460 WQPRYSEYKTSLDINHGQFANGQPLSYWTVGRGRA------GETLETFDIAS---LKINP 510
Query 598 NTLDDLFAVKVDSTWETDQLLVNCNVGCYVTRPLSADGVP 637
LD +FAV + T TD + C +S +G P
Sbjct 511 KWLDSIFAVNYNGTQITDCVFGGCQFNVQKVSDMSENGEP 550
>gi|494306153|ref|WP_007173049.1| hypothetical protein [Prevotella bergensis]
gi|270333881|gb|EFA44667.1| putative capsid protein (F protein) [Prevotella bergensis DSM
17361]
Length=519
Score = 176 bits (447), Expect = 1e-44, Method: Compositional matrix adjust.
Identities = 172/618 (28%), Positives = 259/618 (42%), Gaps = 116/618 (19%)
Query 34 LLPVWTDFAIPNCTYRINLEYFTRTRPVQTSAYTRIREYFDFFAVPCDLIWKSFDSAVIQ 93
LLPV IP+ IN + F RT P+ T+A+ +R ++FF VP +W FD +
Sbjct 2 LLPVLNLDLIPHDHVEINAQDFMRTLPMNTAAFASMRGVYEFFFVPYHQLWAQFDQFITG 61
Query 94 MGEKAPIQSKDLLTNLTVKGDLPYCSLSDLSFALKFASGNPASLGNKVS--VSPGFGNIF 151
M + +K + T +PY +L + N + +P F +
Sbjct 62 MNDFHSSANKSIQGG-TSPLQVPYFNLESV-------------FKNIIERDSTPSFQDDL 107
Query 152 GYNRGDVNHKLLSMLDYGNVVDKSANWIGTNANRWWNRRVATADSAVYSQKYNVNLAVNL 211
Y +LL +L YG D N V K N++ ++
Sbjct 108 QYRFKYGAFRLLDLLGYGRKFDSFGTAYPDN---------------VSGLKNNLDYNCSV 152
Query 212 FFLATYQKIYQDFFRWSQWENADPTSYNFDWYRGSGNLFGSSDLAAAIPASSDYWKRDNL 271
F + Y KIYQD++R S +EN D S+NFD ++G G D A + A +L
Sbjct 153 FRVLAYNKIYQDYYRNSNYENFDTDSFNFDKFKG-----GLVD--AKVVA--------DL 197
Query 272 FSLRYANWNKDKFMGILPNSQFGDLAVVDLGTISASGSKIPVGAYDGVNDSGTFKQFQTS 331
F LRY N D F + + F + IP + D + F + Q +
Sbjct 198 FKLRYRNAQTDYFTNLRQSQLF---------------TFIPEFSDD---EHLNFDRDQYA 239
Query 332 SESYNGSSDKTTQIYPRNPDSISVRADSNLWAVLGESSDLKLKFSVLALRQAEALQKWKE 391
+S S+ T +P V D+NL FSV +LR A A+ K
Sbjct 240 DQS---KSNFTQLNFP-------VDVDNNLGY-----------FSVSSLRSAFAVDKLLS 278
Query 392 ITQSVDTNYRDQIKAHFGVSVPQSEAHMAKYIGGVARNLDISEVVN-------NFLPGPG 444
+T ++DQ++AH+GV +P S Y+GG +L +S+V + P G
Sbjct 279 VTMRAGKTFQDQMRAHYGVEIPDSRDGRVNYLGGFDSDLQVSDVTQTSGTTATEYKPEAG 338
Query 445 NDSSQAYIYGKGVGSGQGSMTFNTGSGYYILMCIYHAVPLLDYSISGPDGQNLVTSVEDL 504
I GKG GSG+G + F+ + +LMCIY VP + Y + D D
Sbjct 339 ---YLGRIAGKGTGSGRGRIVFDAKE-HGVLMCIYSLVPQIQYDCTRLDPMVDKLDRFDF 394
Query 505 PIPEFDNIGMESVPAVELMNSSLYSNVNSAD---KILGYSPRYYNWKTKIDRIHGAFTT- 560
PEF+N+GM+ +NSS S+ + D +LGY PRY +KT +D HG F
Sbjct 395 FTPEFENLGMQP------LNSSYISSFCTPDPKNPVLGYQPRYSEYKTALDINHGQFAQN 448
Query 561 -TLKDWVAPIDDSFLYSLFGDSLSTYKGVTWPFFKVNPNTLDDLFAVKVDSTWETDQLLV 619
L W S F +T+ + FK++P L+ +F V+ + T TD +
Sbjct 449 DALSSWSV--------SRFR-RWTTFPQLEIADFKIDPGCLNSVFPVEFNGTESTDCVFG 499
Query 620 NCNVGCYVTRPLSADGVP 637
CN +S DG+P
Sbjct 500 GCNFNIVKVSDMSVDGMP 517
Lambda K H a alpha
0.318 0.134 0.414 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 4813850017872