bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-5_CDS_annotation_glimmer3.pl_2_3
Length=587
Score E
Sequences producing significant alignments: (Bits) Value
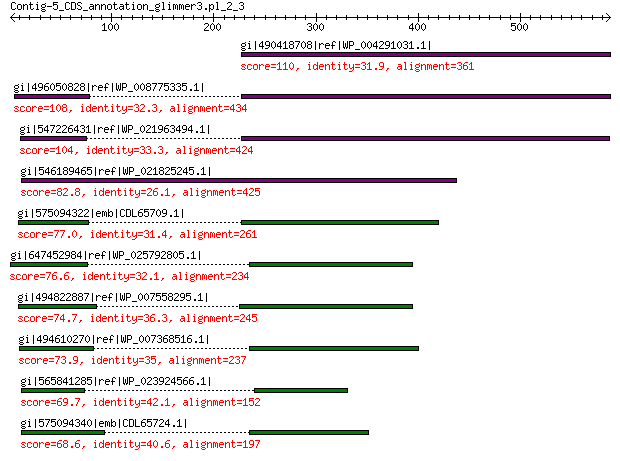
gi|490418708|ref|WP_004291031.1| hypothetical protein 110 9e-23
gi|496050828|ref|WP_008775335.1| hypothetical protein 108 1e-21
gi|547226431|ref|WP_021963494.1| predicted protein 104 2e-20
gi|546189465|ref|WP_021825245.1| hypothetical protein 82.8 2e-13
gi|575094322|emb|CDL65709.1| unnamed protein product 77.0 1e-11
gi|647452984|ref|WP_025792805.1| hypothetical protein 76.6 1e-11
gi|494822887|ref|WP_007558295.1| hypothetical protein 74.7 7e-11
gi|494610270|ref|WP_007368516.1| hypothetical protein 73.9 1e-10
gi|565841285|ref|WP_023924566.1| hypothetical protein 69.7 2e-09
gi|575094340|emb|CDL65724.1| unnamed protein product 68.6 5e-09
>gi|490418708|ref|WP_004291031.1| hypothetical protein [Bacteroides eggerthii]
gi|217986635|gb|EEC52969.1| hypothetical protein BACEGG_02720 [Bacteroides eggerthii DSM
20697]
Length=422
Score = 110 bits (275), Expect = 9e-23, Method: Compositional matrix adjust.
Identities = 115/398 (29%), Positives = 177/398 (44%), Gaps = 52/398 (13%)
Query 227 GLLKYINYRDYQLFAKRFRKYLFTRIGSYEKISSYVVSEYSPKTFRPHFHILFFFDSDEV 286
G L Y+ D QLF KRFR Y+ R EK+ + + EY P FRPH+HIL F SDE
Sbjct 39 GYLPYLRKFDLQLFFKRFRYYVAKRFPK-EKVRYFAIGEYGPVHFRPHYHILLFLQSDEA 97
Query 287 AKNIRQAVFQSWKLGRVDTQLARDSAGSYVSGYLNSVVSLPGIFTDVS---FTKNKSRFS 343
+ + V ++W GRVD QL++ SYV+GY+NS V +P + T + F + +
Sbjct 98 LQVCSKVVSEAWPFGRVDCQLSKGKCSSYVAGYVNSSVLVPKVLTLPTLCPFCVHSQKLG 157
Query 344 KLFGYESFRQTIK--TPEQAIERLSERILFVRNGKPCEFTPPFSYISRLLPRFVPYSSNF 401
+ F +S R + TPEQ ++R V NG+ EF S + P+ F
Sbjct 158 QGF-LQSERAKVYSLTPEQFVKR-----SIVINGRYKEFDVWRSAYAYFFPK----CKGF 207
Query 402 VVETRTVLRSIRGVLQLFRRNEPFKKETPTNISEFVHTYTVTLYEKYGYCYDTLPEC--- 458
++ G+ RR P + T E V +K YC D E
Sbjct 208 ADKSSRERAYSYGLYDTARRLFPSAETTFALAKEIVGYIYYFHNKKDTYCLDIFGEVSDQ 267
Query 459 --LRVYLSYTRSVKEIHYFTDKLKNKLCRPLYIYRIWESLGLSDDCI------------- 503
L + Y + ++Y D + ++CR Y++R++ L LS +
Sbjct 268 SDLYQFSQYFFEPEIVNYSLDSI--EMCR--YVHRVYTELLLSKHFLYFVCDRPTLSEQK 323
Query 504 --ISLSDEYMSRCRSMSLEKQLSIQQEMFEREGYSD-ELLSLFYINKPQKKV-NNRYFQE 559
+ L +E+ SR M L+ QQ +E + D +L+S + N +N YF
Sbjct 324 RKLKLIEEFYSRLDYMHLKTFFENQQLFYESDLVGDLDLMSDAWENSYYPFFYDNVYFSS 383
Query 560 --WKDKNYYEVHYI--------RVKHKKLNDENNVFLE 587
+K Y ++ + R+KHKKLND N +F++
Sbjct 384 EVYKKTPVYRLYDMQISKLFSDRIKHKKLNDLNKIFVD 421
>gi|496050828|ref|WP_008775335.1| hypothetical protein [Bacteroides sp. 2_2_4]
gi|229448892|gb|EEO54683.1| hypothetical protein BSCG_01608 [Bacteroides sp. 2_2_4]
Length=497
Score = 108 bits (269), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 113/406 (28%), Positives = 169/406 (42%), Gaps = 68/406 (17%)
Query 227 GLLKYINYRDYQLFAKRFRKYLFTRIGSYEKISSYVVSEYSPKTFRPHFHILFFFDSDEV 286
G + Y+ D QLF KR R Y+ T+ EK+ + V EY P FRPH+H+L F SDE
Sbjct 115 GDVPYLRKTDLQLFLKRLRYYV-TKQKPSEKVRYFAVGEYGPVHFRPHYHLLLFLQSDEA 173
Query 287 AKNIRQAVFQSWKLGRVDTQLARDSAGSYVSGYLNSVVSLPGIFTDVS---FTKNKSRFS 343
+ + + ++W GRVD Q+++ +YV+ Y+NS ++P +F S F+ + +
Sbjct 174 LQICSENISKAWTFGRVDCQVSKGQCSNYVASYVNSSCTIPKVFKASSVCPFSVHSQKLG 233
Query 344 KLFGYESFRQTI--KTPEQAIERLSERILFVRNGKPCEFTPPFSYISRLLPR---FVPYS 398
+ F + R+ I TPE I R V NGK EF S S PR FV S
Sbjct 234 QGF-LDCQREKIYSLTPENFI-----RSSIVLNGKYKEFDVWRSCYSFFYPRCKGFVTKS 287
Query 399 SNFVVETRTVLRSIRGVLQLFRRNEPFKKETPTNISEFVHTYTVTLYEKYGYCYDTLPEC 458
S + ++ + R + + KE I F + L + YGYC D
Sbjct 288 SRERAYSYSIYDTARLLFPDAKTTFSLAKEIAIYIYYFHNPKETYLLDLYGYCSDQ---- 343
Query 459 LRVYLSYTRSVKEIHYFTDKLKNKLCR---PLYIYRIWESLGLSDDCII----------- 504
S + + Y +D L + Y++RI+ L +S +
Sbjct 344 -----SKLYELSQYFYDSDVLLHSFNSGEFSRYVHRIYTELLISKHFLYFVCTHNTLAER 398
Query 505 ----SLSDEYMSRCRSMSLEKQLSIQQEMFEREGYSDELLSLFYINKPQKKVNNRYF--- 557
L +E+ SR M L K QQ +E + D+ L +N Y+
Sbjct 399 KSKQRLIEEFYSRLDYMHLTKFFEAQQLFYESDLIGDDDLC-------TDNWDNSYYPYF 451
Query 558 --QEWKDKNYYEVHYI--------------RVKHKKLNDENNVFLE 587
+ D N +E + R+KHKKLND N VF E
Sbjct 452 YNNVYTDTNLFEKTPVYRLYSSDVKKLFNDRIKHKKLNDANKVFFE 497
Score = 61.6 bits (148), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 27/73 (37%), Positives = 43/73 (59%), Gaps = 0/73 (0%)
Query 5 VRNFFNRCEHPRIIKNKYTGEPVYVECGSCPHCLISRSDAKRNLCDYEKWNRKYCYFVTL 64
V+N F +C HP+ I N YT E + V CG C C ++++ CD E + K+ F+TL
Sbjct 2 VQNPFCKCLHPKRIMNPYTKESMVVPCGHCQACTLAKNSRYAFQCDLESYTAKHTLFITL 61
Query 65 TYNSQYVPKMALV 77
TY ++++P+ V
Sbjct 62 TYANRFIPRAMFV 74
>gi|547226431|ref|WP_021963494.1| predicted protein [Prevotella sp. CAG:1185]
gi|524103383|emb|CCY83995.1| predicted protein [Prevotella sp. CAG:1185]
Length=498
Score = 104 bits (259), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 108/388 (28%), Positives = 170/388 (44%), Gaps = 46/388 (12%)
Query 227 GLLKYINYRDYQLFAKRFRKYLFTRIGSYEKISSYVVSEYSPKTFRPHFHILFFFDSDEV 286
G L Y + RD QLF KR RK L S EKI Y+VSEY PKTFR H+H+LFF+D +
Sbjct 128 GYLSYTSKRDAQLFLKRVRKNLSKY--SDEKIRYYIVSEYGPKTFRAHYHVLFFYDEVKT 185
Query 287 AKNIRQAVFQSWKLGRVDTQLARDSAGSYVSGYLNSVVSLPGIFTDVS---FTKNKSRFS 343
K + + + Q+W+ GRVD L+R SYV+ Y+N LP D+S F+ + RF+
Sbjct 186 QKVMSKVIRQAWQFGRVDCSLSRGKCNSYVARYVNCNYCLPRFLGDMSTKPFSCHSIRFA 245
Query 344 KLFGYESFRQTIKTPEQAIERLSERILFVRNGKPCEFTPPFSYISRLLPRFVPY---SSN 400
+ K S I NG EF P + P+ Y S +
Sbjct 246 LGIHQSQKEEIYKGSVDDFIYQSGEI----NGNYVEFMPWRNLSCTFFPKCKGYSRKSDS 301
Query 401 FVVETRTVLRSIRGVLQLFRRNEPFKKETPTNISEFVHTYTVTLYEKYGYCYDTLPECLR 460
+ ++ +LR +R + + N + I + V T + + C + P +
Sbjct 302 ELWQSYNILREVRSAIG-YSFNTII--DYARCILDLVVTAKFSCDSRGLPC--SSPALNK 356
Query 461 VYLSYTRSVKEIHYFTDKLK----NKLCRPLYIYRIWESLGLSDDCIISLSDEYMSRCRS 516
V +++ + Y++D L N + R LYI R + + + +D Y R R
Sbjct 357 VISYFSQGIDTNPYYSDYLADYHTNSIARELYISRHFLTF-------VCDNDSYHERYRK 409
Query 517 MSLEKQL------SIQQEMFEREGYSDELLS---LFYINK-PQKKVNNRYFQEWKDKNYY 566
+L +Q ++ M+ + + L+S +YINK P N + + +Y
Sbjct 410 FTLIRQFWQRYDYALLVGMYTSQIENRHLISNYDWYYINKTPLDSFGNVDVSQLSKELFY 469
Query 567 EVHYIR--------VKHKKLNDENNVFL 586
+ I+ +KHK ND N F+
Sbjct 470 KRFVIKSDENFEKSIKHKIQNDANGFFI 497
Score = 74.7 bits (182), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 33/64 (52%), Positives = 43/64 (67%), Gaps = 0/64 (0%)
Query 11 RCEHPRIIKNKYTGEPVYVECGSCPHCLISRSDAKRNLCDYEKWNRKYCYFVTLTYNSQY 70
+C HPR ++NKYTGE + V CG C CL R+D LC E+ + KYC F TLTY++ Y
Sbjct 10 KCYHPRHVQNKYTGEVIQVGCGVCKACLKRRADKMSFLCAIEEQSHKYCMFATLTYSNDY 69
Query 71 VPKM 74
VP+M
Sbjct 70 VPRM 73
>gi|546189465|ref|WP_021825245.1| hypothetical protein [Prevotella salivae]
gi|544001993|gb|ERK01417.1| hypothetical protein HMPREF9145_2741 [Prevotella salivae F0493]
Length=586
Score = 82.8 bits (203), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 111/452 (25%), Positives = 183/452 (40%), Gaps = 96/452 (21%)
Query 12 CEHPRIIKNKYTGEPVYVECGSCPHCLISRSDAKRNLCDYEKWNRKYCYFVTLTYNSQYV 71
C P I++N G V CG C CL ++ R E + K+C F TLTY++++V
Sbjct 17 CTSPIIVEN--NGRKYAVACGKCECCLHKKATIWRTRLRQEMKDNKFCLFFTLTYDNEHV 74
Query 72 PKMALVPITD-YEFDYPIGKNWPCVKTQLHMRLMLDPRVKKTEMFSQETRMRARRPVGDS 130
P D Y D G V+ + L P+ R V S
Sbjct 75 PFFGRAKNNDFYTLDGEEG-----VQLKGDDNLTYSPK----------------RSVPTS 113
Query 131 LINMFTCKVNKPYIDEHLKAIFDSCAAAVEFRKNYKPTYQSPARPYILRTIPRISKLQKF 190
L C+ P I FD C + + R+ QKF
Sbjct 114 L-----CRDGVPTITN-----FDVCDS--------------------FAVVSRVDA-QKF 142
Query 191 NDVQREELVWLSPEKVEMLKKK-----SKCEGDNNAFPQFKGLLKYINYRDYQLFAKRFR 245
R L L + +++ + ++ G + + P FK L ++ Y L+ ++
Sbjct 143 MKRFRWHLFHLLVKHYKLIFQDKLFTFTQYLGYDGSIP-FKEWLDDLDTETYDLYYSVYQ 201
Query 246 KYLFTRIGSYEK--------ISSYVVSEYSPKTFRPHFHILFFFDSDEVAKNIRQAVFQS 297
YL YEK + ++ SEY+P TFRPHFH LF+FD ++ + +F++
Sbjct 202 YYL----TDYEKKKESCKQSVRYFICSEYTPTTFRPHFHGLFWFDDEKAFSYAPRCIFKA 257
Query 298 WKLG---RVDTQLARDSAGSYVSGYLNSVVSLPGIFTDVSFTKNKSRFSK--LFGYESFR 352
WK+ ++ Q A +YVS Y+ +LP + S T+ SK GY+SF
Sbjct 258 WKMCAEININVQPVSGDASAYVSKYVTGNSNLPPVLQAKS-TRTFCLASKGPAIGYKSF- 315
Query 353 QTIKTPEQAIERLSERILF------VRNGKPCEFTP-PFSYISRLLPRFVPYSS-NFVVE 404
+ ++ +E + R +F + GK + P S + R P+ YS+ ++V +
Sbjct 316 ----SDKEVLEMFTRRCIFRSYETISKKGKLSGVSAVPSSAVGRYFPKCYQYSTLSYVDK 371
Query 405 TRTVLRSIRGVLQLFRRNEPFKKETPTNISEF 436
R R I+ L + + K + P +++ F
Sbjct 372 FRVYTRFIK----LSKHDGVEKIDIPASLACF 399
>gi|575094322|emb|CDL65709.1| unnamed protein product [uncultured bacterium]
Length=499
Score = 77.0 bits (188), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 61/210 (29%), Positives = 92/210 (44%), Gaps = 21/210 (10%)
Query 227 GLLKYINYRDYQLFAKRFRKYLFTRIGSYEKISSYVVSEYSPKTFRPHFHILFFFDSDEV 286
GL + YRD QLF KR RK+++ G EKI Y++ EY K+ RPH+H L FF+S +
Sbjct 137 GLYALLYYRDIQLFLKRLRKHIYKYYG--EKIRFYIIGEYGTKSLRPHWHCLLFFNSSSL 194
Query 287 AKNIRQAVFQS---------------WKLGRVDTQLARDSAGSYVSGYLNSVVSLPGIFT 331
++ V W+ G D++ A +YVS Y+N + P +
Sbjct 195 SQAFEDCVNVGTTSRPCSCPRFLRPFWQFGICDSKRTNGEAYNYVSSYVNQSANFPKLL- 253
Query 332 DVSFTKNKSRFSKLFGYESFRQTIKTPEQAIE-RLSERILFVRN-GKPCEFTPPFSYISR 389
V + K+ S G Q+I + Q + ER ++ G ++ SY SR
Sbjct 254 -VLLSNQKAYHSIQLGQILSEQSIVSAIQKGDFSFFERQFYLDTFGAANSYSVWRSYYSR 312
Query 390 LLPRFVPYSSNFVVETRTVLRSIRGVLQLF 419
P+F S +T VL + LF
Sbjct 313 FFPKFTCSSQLTYEQTYRVLTCYETLRDLF 342
Score = 40.8 bits (94), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 21/68 (31%), Positives = 36/68 (53%), Gaps = 1/68 (1%)
Query 9 FNRCEHPRIIKNKYTGEPVYVECGSCPHCLISRSDAKRNLCDYEKWNRKYCYFVTLTYNS 68
F +C P ++++ G P V CG C C ++ + E++ KYCYF+TLTY+
Sbjct 5 FVKCFSPLVLRDP-RGYPYQVPCGKCIACHNNKRSSLSLKLRLEEYTSKYCYFLTLTYDD 63
Query 69 QYVPKMAL 76
+P ++
Sbjct 64 DNLPLFSV 71
>gi|647452984|ref|WP_025792805.1| hypothetical protein [Prevotella histicola]
Length=480
Score = 76.6 bits (187), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 50/171 (29%), Positives = 80/171 (47%), Gaps = 20/171 (12%)
Query 235 RDYQLFAKRFRKYLFTRI---GSYEKISSYVVSEYSPKTFRPHFHILFFFDSDEVAKNIR 291
+D Q F KR R + ++ G+ +I ++ SEY P TFRPH+H + ++DS+ + +
Sbjct 125 KDVQDFFKRLRSKIDYKLKPRGNEYRIRYFICSEYGPNTFRPHYHAILWYDSEILHNELN 184
Query 292 QAVFQSWKLGRVDTQLARDSAGSYVSGYLNSVVSLPGIFTDVSFTKNKSRFSKLFGYESF 351
+ ++WK G D L SA YV+ Y+N LP ++ F+ F S
Sbjct 185 VLIRETWKNGNTDFSLVNSSASQYVAKYVNGDCDLPSFL--------RTEFTSTFHLASK 236
Query 352 RQTIKTPEQAIERLSERILFVRNGKPC--------EFT-PPFSYISRLLPR 393
I + E L E ++ G+ C EF PP S +R+LP+
Sbjct 237 HPCIGYGKDDEEALYENVINGTYGRNCLNKSTNEFEFVCPPRSLENRILPK 287
Score = 43.5 bits (101), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 25/75 (33%), Positives = 36/75 (48%), Gaps = 0/75 (0%)
Query 1 MADKVRNFFNRCEHPRIIKNKYTGEPVYVECGSCPHCLISRSDAKRNLCDYEKWNRKYCY 60
M + N C P I N+Y GE +Y C C C S + + N D E +Y
Sbjct 1 MFSRALGNINPCLRPHRIYNRYIGEFLYTNCRKCVRCRSSYASSWANRIDSECSFHRYSL 60
Query 61 FVTLTYNSQYVPKMA 75
F+TLTY++ ++P A
Sbjct 61 FLTLTYDNDHLPYYA 75
>gi|494822887|ref|WP_007558295.1| hypothetical protein [Bacteroides plebeius]
gi|198272100|gb|EDY96369.1| hypothetical protein BACPLE_00805 [Bacteroides plebeius DSM 17135]
Length=545
Score = 74.7 bits (182), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 59/200 (30%), Positives = 86/200 (43%), Gaps = 35/200 (18%)
Query 225 FKGLLKYINYRDYQLFAKRFRKYLFTRIGSYEKISSYVVSEYSPKTFRPHFHILFFFDS- 283
+KG Y++ R+ QLF KR RKYL G +KI + EY P +FRPHFHIL F D
Sbjct 114 YKGRFPYLSKRELQLFMKRLRKYLDKYEG--QKIRFFATGEYGPLSFRPHFHILLFVDDP 171
Query 284 ----------------------------DEVAKNIRQAVFQSWKLGRVDTQ-LARDSAGS 314
+ + + +SW G +D Q + + S S
Sbjct 172 SLFLPSVHTLGEYPYPYWSKYQKAHCGKGTLLSKLEYYIRESWPFGGIDAQSVEQGSCSS 231
Query 315 YVSGYLNSVVSLPGIFTDVSFTKNKSRFSKLFGYESF-RQTIKTPEQAIERLSERILFVR 373
YV+GY+NS V LP V K+ S+ S+ G + F + I + +R F R
Sbjct 232 YVAGYVNSSVPLPSCLK-VDAVKSFSQHSRFLGRKIFGTELIPLLKLKFTEFVQRSFFCR 290
Query 374 NGKPCEFTPPFSYISRLLPR 393
G+ F P + + P+
Sbjct 291 -GRYDNFRTPSEMLHSVYPQ 309
Score = 60.8 bits (146), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 30/76 (39%), Positives = 42/76 (55%), Gaps = 0/76 (0%)
Query 9 FNRCEHPRIIKNKYTGEPVYVECGSCPHCLISRSDAKRNLCDYEKWNRKYCYFVTLTYNS 68
F C P+ IKNKYTGE + V C C C R+ NLCD+E K F+TLT++
Sbjct 5 FVSCLEPQRIKNKYTGEEMVVACKHCVACEQLRNFKYSNLCDFESLTAKKTVFLTLTFDD 64
Query 69 QYVPKMALVPITDYEF 84
++VP+ + D E+
Sbjct 65 KFVPQFRFYKVGDDEY 80
>gi|494610270|ref|WP_007368516.1| hypothetical protein [Prevotella multiformis]
gi|324988542|gb|EGC20505.1| hypothetical protein HMPREF9141_0984 [Prevotella multiformis
DSM 16608]
Length=479
Score = 73.9 bits (180), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 60/178 (34%), Positives = 88/178 (49%), Gaps = 21/178 (12%)
Query 235 RDYQLFAKRFRK---YLFTRIGSYE-KISSYVVSEYSPKTFRPHFHILFFFDSDEVAKNI 290
+D Q + KR R Y + S E +I ++ SEY P+TFRPH+H + ++DS+E+ +NI
Sbjct 122 KDVQDWFKRLRSAVDYQLNKNKSNEFRIRYFICSEYGPRTFRPHYHAILWYDSEELQRNI 181
Query 291 RQAVFQSWKLGRVDTQLARDSAGSYVSGYLNSVVSLPGIFTDVSFTKNKSRFSK--LFGY 348
+ + ++WK G L +SA YV+ Y+N LP F FT SK GY
Sbjct 182 GRLIRETWKNGNSVFSLVNNSASQYVAKYVNGDTRLPP-FLRTEFTSTFHLASKHPYIGY 240
Query 349 ESFRQTIKTPEQAIER------LSERILFVRNGKPCEFTP-PFSYISRLLPRFVPYSS 399
K E+A+ + +L NG+ EF P P S +RLLP+ Y S
Sbjct 241 ------CKADEEALRSNVLDGTYGQSVLNRDNGQ-FEFVPTPRSLENRLLPKCRGYRS 291
Score = 42.0 bits (97), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 23/72 (32%), Positives = 36/72 (50%), Gaps = 0/72 (0%)
Query 10 NRCEHPRIIKNKYTGEPVYVECGSCPHCLISRSDAKRNLCDYEKWNRKYCYFVTLTYNSQ 69
N C P+ I NKY E +YV C C C S + + E ++ FVTLTY+++
Sbjct 8 NSCLSPKRIYNKYIDETLYVPCRKCFRCRDSYASDWSRRIENECREHRFSLFVTLTYDNE 67
Query 70 YVPKMALVPITD 81
++P + + D
Sbjct 68 HIPLFQPLVMDD 79
>gi|565841285|ref|WP_023924566.1| hypothetical protein [Prevotella nigrescens]
gi|564729906|gb|ETD29850.1| hypothetical protein HMPREF1173_00032 [Prevotella nigrescens
CC14M]
Length=484
Score = 69.7 bits (169), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 39/96 (41%), Positives = 54/96 (56%), Gaps = 5/96 (5%)
Query 240 FAKRFRK---YLFTR--IGSYEKISSYVVSEYSPKTFRPHFHILFFFDSDEVAKNIRQAV 294
F KR R Y F + I + EKI +V SEY PKT RPH+H + +FDS+EVA+ I + +
Sbjct 123 FFKRLRSKLSYYFKKHHIITNEKIRYFVCSEYGPKTLRPHYHAIIWFDSEEVARVIEKML 182
Query 295 FQSWKLGRVDTQLARDSAGSYVSGYLNSVVSLPGIF 330
SW G D + +A YV+ Y++ LP I
Sbjct 183 SSSWSNGFTDFEYVNSTAPQYVAKYVSGNSVLPEIL 218
Score = 46.6 bits (109), Expect = 0.041, Method: Compositional matrix adjust.
Identities = 25/61 (41%), Positives = 34/61 (56%), Gaps = 0/61 (0%)
Query 12 CEHPRIIKNKYTGEPVYVECGSCPHCLISRSDAKRNLCDYEKWNRKYCYFVTLTYNSQYV 71
CEHP+ I N YT E V+V C C CL ++ A E Y FVTLTY+++++
Sbjct 9 CEHPKRIINPYTHERVWVACRRCKCCLNKKTSAWSGRVANECKLHAYSAFVTLTYDNEHL 68
Query 72 P 72
P
Sbjct 69 P 69
>gi|575094340|emb|CDL65724.1| unnamed protein product [uncultured bacterium]
Length=486
Score = 68.6 bits (166), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 49/123 (40%), Positives = 72/123 (59%), Gaps = 10/123 (8%)
Query 235 RDYQLFAKRFRKYLFTRIGSYE-KISSYVVSEYSPKTFRPHFHILFFFDSDEVA-KNIRQ 292
+D+ F KR R L TR +YE KI+ + SEY P T RPHFH +F+FDS ++ + R
Sbjct 157 KDFVNFVKRLRINL-TRNYNYEGKITYFKCSEYGPTTNRPHFHGIFWFDSRALSFDSFRS 215
Query 293 AVFQSWKLGRVDTQ-----LARDSAGSYVSGYLNSVVSLPGIFTDVSFTKNKSRFSKLFG 347
AV +SWK+ D Q +AR+ A +YV+ Y+N + S+P +F + K SK FG
Sbjct 216 AVVESWKMCDKDKQYENVEIAREPA-TYVASYVNCLTSVPPLFL-FKGLRPKHSHSKGFG 273
Query 348 YES 350
+ +
Sbjct 274 FAN 276
Score = 51.6 bits (122), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 31/84 (37%), Positives = 49/84 (58%), Gaps = 8/84 (10%)
Query 12 CEHPRIIKNKYTGEPVYVECGSCPHCLISRSDAKRNLCD-YEKWNRKYCY--FVTLTYNS 68
C + + NKY G YV+CG CP CL + A ++ C ++ R Y + FVTLTY++
Sbjct 6 CTNRIKVTNKYVGRSFYVDCGHCPSCL--QRKANKSCCKIINEYGRPYSFMCFVTLTYDN 63
Query 69 QYVPKMALVPITDYEFDYPIGKNW 92
+++P + P TDY Y +GK++
Sbjct 64 EHIPYIH--PDTDYSHLY-VGKSY 84
Lambda K H a alpha
0.323 0.138 0.422 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 4336743713901