bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-5_CDS_annotation_glimmer3.pl_2_7
Length=378
Score E
Sequences producing significant alignments: (Bits) Value
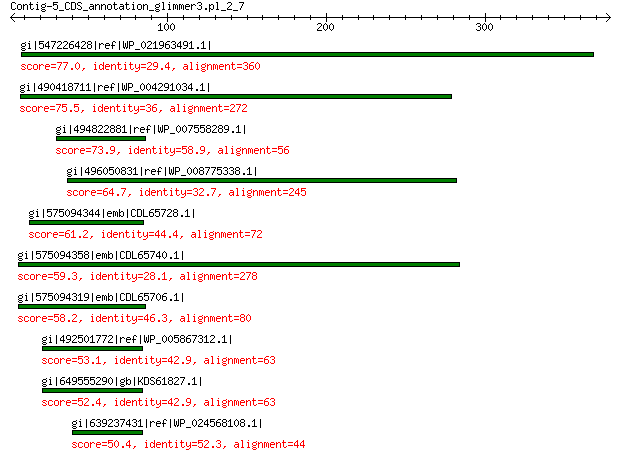
gi|547226428|ref|WP_021963491.1| putative uncharacterized protein 77.0 4e-12
gi|490418711|ref|WP_004291034.1| hypothetical protein 75.5 7e-12
gi|494822881|ref|WP_007558289.1| hypothetical protein 73.9 2e-11
gi|496050831|ref|WP_008775338.1| predicted protein 64.7 3e-08
gi|575094344|emb|CDL65728.1| unnamed protein product 61.2 4e-07
gi|575094358|emb|CDL65740.1| unnamed protein product 59.3 1e-06
gi|575094319|emb|CDL65706.1| unnamed protein product 58.2 4e-06
gi|492501772|ref|WP_005867312.1| hypothetical protein 53.1 1e-04
gi|649555290|gb|KDS61827.1| hypothetical protein M095_3809 52.4 2e-04
gi|639237431|ref|WP_024568108.1| hypothetical protein 50.4 8e-04
>gi|547226428|ref|WP_021963491.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
gi|524103380|emb|CCY83991.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
Length=416
Score = 77.0 bits (188), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 106/417 (25%), Positives = 170/417 (41%), Gaps = 75/417 (18%)
Query 8 SGLFGGLGSVISGAIGAKTTADTNKTNLKIAQMNNDFNAREAQKARDFQLDMWNKENE-- 65
S L G S + G+K+ +DTNKTNL+IAQMNN++N R K ++ DM+N++ E
Sbjct 7 SALIGAGASFLGNIFGSKSQSDTNKTNLQIAQMNNEYNERMFNKQLEYNQDMFNQQVEYD 66
Query 66 ------------------------YNSASSQRERLENAGYNPYM------NEaqagtatg 95
YNSA +QR RLE AG NPY+ A + +
Sbjct 67 QKKMEQQNNFNARMQNEAIGAQQVYNSAKAQRARLEAAGLNPYLMMSGGNAGAVSAVSGS 126
Query 96 msgtsaasaagaaPQI-------PYTPDFQSV--------------GVNLASALKMMSEK 134
+ S G P + PDF V GV A A + +
Sbjct 127 SGSGGSPSPMGVNPPTASSAVMQAFRPDFSGVTGIIQTLLDIQAQKGVRDAQAFSLGEQA 186
Query 135 KKTDIENLNMSDLLRSQIWQNLGATDWRNASPEARAYNLSQGRRAAELG--MASLEENLS 192
IEN ++ L I+ + + +N+ N+S R A ++ +
Sbjct 187 SGFKIENKYKAEKLLWDIYNSKADYNLKNSQESLN--NMSFARLQAMFSSDVSKAQREAE 244
Query 193 NQRWSNNLLVANIANSLLDADTKTILNKYLDQQQQAELNVKAANYEYLVMSGQLKRQEVN 252
N +++ L+ A A L KY DQ+ EL + +A LV +G+ +
Sbjct 245 NAQFTGELIRAQTACQQLQGLLGAKELKYYDQKVLQELAIMSAQQYSLVAAGKASEAQAR 304
Query 253 NLIAEEIETYARANGYNLQNRILRETSDGLIR-ATNNTNFYFGSYYHSRAFNAGADAFHD 311
I + + G + N + ++T++ LI+ A NN N SY++S+ H+
Sbjct 305 QAIENALNLVEQREGIKVDNYVKQKTANALIKTARNNCN---TSYWNSK-------TAHN 354
Query 312 SSILRSRAGSASESYKQSAFDTKLQPWREALNSTNMIFNG-IGSGLDSYTNFQNGRY 367
S+ R +S+ Q F+ + + L S IF G +G GL+ Y + R+
Sbjct 355 QSL---RPSVFEDSFSQ-GFNKFINTYIAPLGSA--IFGGAVGYGLNIYNKNSDDRH 405
>gi|490418711|ref|WP_004291034.1| hypothetical protein [Bacteroides eggerthii]
gi|217986638|gb|EEC52972.1| hypothetical protein BACEGG_02723 [Bacteroides eggerthii DSM
20697]
Length=368
Score = 75.5 bits (184), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 98/329 (30%), Positives = 149/329 (45%), Gaps = 63/329 (19%)
Query 7 ASGLFGGLGSVISGAI----GAKTTADTNKTNLKIAQMNNDFNAR--------------- 47
A+ + G +GS I GA TT N+ N +IAQMNN FN +
Sbjct 3 AAAMTGIVGSAIGAGTSLIGGASTTHMQNQANKEIAQMNNAFNEKMFDKQIAYNKEMYQT 62
Query 48 ---------EAQKA---------RDFQLDMWNKENEYNSASSQRERLENAGYNPY--MNE 87
+ QKA + +Q +MWNK+NEYN S+QR RLE AG NPY MN
Sbjct 63 QLGDQWKFYDDQKANAWKLYEDNKAYQTEMWNKQNEYNDPSAQRARLEAAGLNPYMMMNG 122
Query 88 aqagtatgmsgtsaasaagaaPQ---------IPYTPDFQSVGVNLASALKMMSEKKKTD 138
AG A +SGT ++ + +P PY+ D+ V L A+ + + +
Sbjct 123 GSAGVAGSVSGTQGSAPSAGSPSAQGVQPPTATPYSADYSGVMQGLGHAIDTIMTGSQRN 182
Query 139 IENLNMSDLLRSQIWQNLGATDWRNASPEA-RAYNLSQG---RRAAELGMASLEENLS-N 193
I+N +D LR + G A E + YN ++ R A + ++S++++LS +
Sbjct 183 IQNAQ-ADNLRIE-----GKYIASKAIAELYKTYNEAKNDDERVAIQRVLSSIQKDLSAS 236
Query 194 QRWSNNLLVANIANSLLDADTKTILN----KYLDQQQQAELNVKAANYEYLVMSGQLKRQ 249
Q NN V I A T+ +L K+L +Q+ +L + AA+ L +
Sbjct 237 QVAVNNENVRQIQAQTKIAVTENLLREQQLKFLPYEQRTQLALGAADIALKYAQKNLTEK 296
Query 250 EVNNLIAEEIETYARANGYNLQNRILRET 278
+ + I + ET RANG +QN+ ET
Sbjct 297 QARHEIEKLAETIVRANGQAMQNQYDAET 325
>gi|494822881|ref|WP_007558289.1| hypothetical protein [Bacteroides plebeius]
gi|198272097|gb|EDY96366.1| hypothetical protein BACPLE_00802 [Bacteroides plebeius DSM 17135]
Length=344
Score = 73.9 bits (180), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 33/56 (59%), Positives = 45/56 (80%), Gaps = 0/56 (0%)
Query 30 TNKTNLKIAQMNNDFNAREAQKARDFQLDMWNKENEYNSASSQRERLENAGYNPYM 85
TN+ N++IAQM+N++N + ++ + + DMWN ENEYNSASSQR+RLE AG NPYM
Sbjct 43 TNQANIQIAQMSNEYNREQLERQIEQEWDMWNAENEYNSASSQRKRLEEAGLNPYM 98
>gi|496050831|ref|WP_008775338.1| predicted protein [Bacteroides sp. 2_2_4]
gi|229448895|gb|EEO54686.1| hypothetical protein BSCG_01611 [Bacteroides sp. 2_2_4]
Length=381
Score = 64.7 bits (156), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 80/283 (28%), Positives = 125/283 (44%), Gaps = 45/283 (16%)
Query 37 IAQMNNDFNAREAQKARDFQL----------------------DMWNKENEYNSASSQRE 74
IAQMNN+FN R QK D+ DM+N NEYNSAS+QRE
Sbjct 41 IAQMNNEFNERMLQKQMDYNTLAYDQQVSDQWSFYNDAKQNAWDMFNATNEYNSASAQRE 100
Query 75 RLENAGYNPYMNEaqagtatgmsgtsaasaagaaPQI------PYTPDFQSVGVNLASAL 128
R E AG NPY+ T + ++ ++ A I PY+ D+ + L A+
Sbjct 101 RYEAAGLNPYVMMNTGSAGTAAATSATSATAPTKQGITPPTASPYSADYSGIMQGLGQAI 160
Query 129 KMMS---EKKKTDIENLNMSDLLRSQIWQNLGATDWRNASPEARAYNLSQGRRAAELGMA 185
+S +K KT E N+ + + + + R A+ +A ++ + +L M
Sbjct 161 DQLSSIPDKAKTIAETGNLKIEGKYKAAEAIA----RIANIKADTHSKKEQVALNKL-MY 215
Query 186 SLEENLSNQRWSNNLLVANIANSLLDADTKTILN-------KYLDQQQQAELNVKAANYE 238
S++++L++ + N NIAN + K I ++D Q+ EL KAAN +
Sbjct 216 SIQKDLASSTMAVN--SQNIANMRAEEKFKNIQTLIADKQLSFMDATQKMELAEKAANIQ 273
Query 239 YLVMSGQLKRQEVNNLIAEEIETYARANGYNLQNRILRETSDG 281
+ G L R + + I + ET AR N Q + E + G
Sbjct 274 LKLAQGALTRNQAAHEIKKISETEARTTLINEQTSLTIEQNTG 316
>gi|575094344|emb|CDL65728.1| unnamed protein product [uncultured bacterium]
Length=368
Score = 61.2 bits (147), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 32/72 (44%), Positives = 44/72 (61%), Gaps = 5/72 (7%)
Query 13 GLGSVISGAIGAKTTADTNKTNLKIAQMNNDFNAREAQKARDFQLDMWNKENEYNSASSQ 72
GLG ++ G G ++ + QM ++ ++EAQK RDFQLDMWN+ NEYN Q
Sbjct 26 GLG-IVDGVAG--MFGQSSDQRFALGQM--EWQSQEAQKQRDFQLDMWNRNNEYNKPDEQ 80
Query 73 RERLENAGYNPY 84
+RLE AG NP+
Sbjct 81 MKRLEEAGINPW 92
>gi|575094358|emb|CDL65740.1| unnamed protein product [uncultured bacterium]
Length=328
Score = 59.3 bits (142), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 78/300 (26%), Positives = 142/300 (47%), Gaps = 33/300 (11%)
Query 6 FASGLFGGLGSVISGAIGAKTTAD-TNKTNLKIAQMNNDFNAREAQKARDFQLDMWNKEN 64
A G +G+ + AI K D TN TN++IAQMNN+++ R +K + +MW K
Sbjct 1 MAVGTMIDIGAGLYTAISNKNAVDNTNSTNMQIAQMNNEWSERMMEKQMAYNTEMWEKVA 60
Query 65 EYNSASSQRERLENAGYNPYMNEaqagtatgmsgtsaasaagaaPQIPYTP---DFQSV- 120
+YNS ++ ++ +AG NPYM + + + ++ + + + Q+ P DF SV
Sbjct 61 DYNSLPNKMQQARDAGVNPYMALSGNAFGSISAPSANSVSLPSPSQVQAQPAQYDFSSVS 120
Query 121 -----GVNLASALKMMSEKK-----KTD---IENLNMSDLLRSQIWQNLGATDWRNASPE 167
G++L ++M ++ TD IEN + L S+I + + T +
Sbjct 121 NSIIAGMDLFQKAQLMKSQQSNIDASTDQLRIENKYHAMKLVSEIAEKMANTK----DSQ 176
Query 168 ARAYNLSQGRRAAELGMAS-LE---ENLSNQRWSNNLLVANIANSLLDADTKTILNKYLD 223
A+A AE G+ + LE + LSN + + LV L +A T L ++
Sbjct 177 AKAVYQQIINEYAEQGIKTDLEIKNQTLSNMKETFRGLV------LENAMTSEQL-RFFP 229
Query 224 QQQQAELNVKAANYEYLVMSGQLKRQEVNNLIAEEIETYARANGYNLQNRILRETSDGLI 283
+Q +A+L + A+ + +L +Q++ I + +T + G + N ILR+ + ++
Sbjct 230 EQVRAQLGLTASQILLNQSNSKLSQQKMVESIYNQWKTDSERQGIQINNSILRKAAKDIV 289
>gi|575094319|emb|CDL65706.1| unnamed protein product [uncultured bacterium]
Length=396
Score = 58.2 bits (139), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 37/91 (41%), Positives = 46/91 (51%), Gaps = 22/91 (24%)
Query 6 FASGLFGGLGSVISGAI---GAKTTA--------DTNKTNLKIAQMNNDFNAREAQKARD 54
+GL G GS+I+G G+K A +TN+ N +IA NN FN R
Sbjct 24 LGAGLIAGAGSLINGLFSSNGSKQAAKYQLQAVRETNQANREIADQNNKFNER------- 76
Query 55 FQLDMWNKENEYNSASSQRERLENAGYNPYM 85
MWN +NEYN QR RLE AG NPY+
Sbjct 77 ----MWNLQNEYNRPDMQRARLEAAGLNPYL 103
>gi|492501772|ref|WP_005867312.1| hypothetical protein [Parabacteroides distasonis]
gi|409230405|gb|EKN23269.1| hypothetical protein HMPREF1059_03254 [Parabacteroides distasonis
CL09T03C24]
Length=288
Score = 53.1 bits (126), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 27/63 (43%), Positives = 38/63 (60%), Gaps = 0/63 (0%)
Query 21 AIGAKTTADTNKTNLKIAQMNNDFNAREAQKARDFQLDMWNKENEYNSASSQRERLENAG 80
A+ K DTNK N++IA+ + +E +KA L+MWN +NEYNS + Q R+ AG
Sbjct 22 AMNNKAVQDTNKANMEIAKYQAQWQQQENEKAYQRSLNMWNLQNEYNSPTQQMARIRAAG 81
Query 81 YNP 83
NP
Sbjct 82 LNP 84
>gi|649555290|gb|KDS61827.1| hypothetical protein M095_3809 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649557306|gb|KDS63785.1| hypothetical protein M095_3404 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649559158|gb|KDS65545.1| hypothetical protein M096_4689 [Parabacteroides distasonis str.
3999B T(B) 6]
gi|649560567|gb|KDS66875.1| hypothetical protein M095_2448 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649561016|gb|KDS67303.1| hypothetical protein M095_2410 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649562727|gb|KDS68911.1| hypothetical protein M096_3341 [Parabacteroides distasonis str.
3999B T(B) 6]
Length=288
Score = 52.4 bits (124), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 27/63 (43%), Positives = 37/63 (59%), Gaps = 0/63 (0%)
Query 21 AIGAKTTADTNKTNLKIAQMNNDFNAREAQKARDFQLDMWNKENEYNSASSQRERLENAG 80
A+ K DTNK N++IA+ + +E +KA L MWN +NEYNS + Q R+ AG
Sbjct 22 AMNNKAVQDTNKANMEIAKYQAQWQQQENEKAYQRSLKMWNLQNEYNSPTQQMARIRAAG 81
Query 81 YNP 83
NP
Sbjct 82 LNP 84
>gi|639237431|ref|WP_024568108.1| hypothetical protein [Elizabethkingia anophelis]
Length=287
Score = 50.4 bits (119), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 23/44 (52%), Positives = 31/44 (70%), Gaps = 0/44 (0%)
Query 40 MNNDFNAREAQKARDFQLDMWNKENEYNSASSQRERLENAGYNP 83
MNN N + A++ R F LDMWN+ NEYN+ +Q +RL+ AG NP
Sbjct 18 MNNSSNKKIARENRAFALDMWNRNNEYNTPLAQMQRLKEAGLNP 61
Lambda K H a alpha
0.312 0.127 0.357 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 2349684375798